Issue:January/February 2020
IMAGE-BASED CHARACTERIZATION - Non-Invasive, Quantitative Characterization of Lyophilized Drug Product Using Three-Dimensional X-Ray Microscopy Analytics
ABSTRACT
Lyophilization is a freeze-drying process that removes water from a drug product via sublimation. It is a common and important processing method for pharmaceutical products that are not stable in liquid or frozen state, or when cold supply chain strategies are not applicable. Thorough characterization of drug product quality, such as cake moisture content, API purity, and cake appearance has to be conducted before market release. Cake appearance can be a simple way to elucidate critical product defects, but visual inspection may not detect impactful changes in the cake microstructure. An innovative image-based analytical method uses 1) high-resolution, non-invasive three-dimensional imaging with X-Ray Microscopy, 2) artificial intelligence-based image processing, and 3) image-based physical property modeling with direct numerical simulations. This new method can not only characterize quantitatively the microstructures of the lyophilized drug samples, but also presents the potential to correlate the microstructures with physical properties to optimize parameters in drug formulation, cycle development, process scale-up, stability control, and administration.
LYOPHILIZED DRUG PRODUCT
To enhance storage stability of biopharmaceutical products, lyophilization is often used.1,2 Lyophilization is a freeze-drying process that removes water from a drug product via sublimation. It is a common and important processing method for pharmaceutical products that are not stable in liquid or frozen state, or when cold supply chain strategies are not applicable. In addition to sterility and endotoxin testing, physical characterization, such as cake moisture content, reconstitution time, integrity, homogeneity, uniformity, potency, and cake appearance, is often employed to confirm drug product quality. Despite decades of commercialization, lyophilization remains to be time-consuming and costly in both development and scale-up phases due to the challenges in cycle development, process optimization, and cake characterization, particularly when troubleshooting, such as understanding inadequate reconstitution times, is necessary.
An innovative image-based analytical method for cake structure analysis uses high-resolution, non-invasive imaging with X-Ray Microscopy (XRM) collecting three-dimensional (3D) volume data from lyophilized drug samples in the vial. It is important that the cake structure is determined in situ within the vial to avoid impacting any measured cake parameters, such as pore size and porosity, due to manual manipulation. Common structural analysis techniques, such as Brunauer-Emmett-Teller (BET) specific surface area testing or Mercury intrusion capillary porosimetry (MICP), typically require the cake is removed from the vial for analysis. The reconstructed 3D dataset is then quantified with an artificial intelligence (AI)-based image processing method. Image-based modeling with direct numerical simulations are conducted to predict multiple physical properties that are otherwise difficult, or impossible, to measure.
This new image to simulation (I2S) method not only characterizes quantitatively the microstructure of the lyophilized drug samples, but also presents potential to correlate the microstructures with physical properties and process parameters. For example, Pisano, et. al had explored mass transfer characterization using XRay micro-computed tomography images.3 Further enhancement of the method could be used to optimize parameters in drug formulation, cycle development, process scale-up, transfer, and stability.
XRM NON-INVASIVE VISUAL INSPECTION
The opaque nature of lyophilized products makes visual inspection of the cake structure difficult. Removal of the cake from container, required by light microscopy and scanning electron microscopy imaging techniques, is undesirable, as the cake product is highly sensitive to humidity and temperature fluctuations. X-Ray micro-computed tomographic (MicroCT) imaging offers an attractive solution, as it can visualize internal cake structures non-invasively.4 Popular MicroCT systems used in the pharmaceutical industry, however, have a few limitations. First, these MicroCT systems, originally designed for small animal imaging, are limited to 5- to 10-μm resolutions, insufficient to elucidate the sub-micron sized cake microstructures. Second, these MicroCT systems only support geometrical magnification, i.e., they can only obtain higher resolution scans by moving the sample closer to the X-Ray source. For larger samples, their dimension limits nearness to the X-Ray source, and ultimately limiting resolution. Cutting the samples smaller is not always desirable, and often impossible for the lyophilized product. Third, popular small animal imaging MicroCT systems are calibrated to handle small animal tissues and organs. They suffer from a severe beam hardening artifact when imaging cake directly inside a glass vial, due to the high-density gradient between the wall of the vial and the cake.
X-Ray Microscopy (XRM), in comparison, removes all of these limitations. Spatial resolutions of 0.5 to 1 μm, or an order of magnitude improvement from small animal MicroCT imaging systems, is routinely achieved. A multi-stage magnification optics design allows a 3D scan focus on an internal sub-volume of the sample digitally, thus, it supports high resolution at constant distance from the X-Ray source. Beam hardening can be mitigated to obtain artifact-free cake microstructure. XRM is traditionally only considered feasible with a synchrotron facility.5 In recent years, however, it has become increasingly available in a laboratory environment and is more accessible to broader pharmaceutical community.
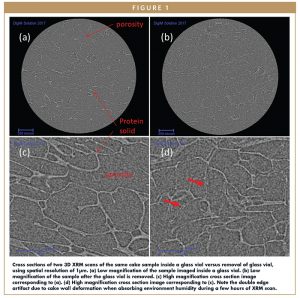
Figure 1 shows two XRM scans of cake sample D342, where the cake microstructures with and without glass vial during the imaging experiments are compared. To better visualize the detail of the microstructure, only one 2D cross section from the 3D volume is shown. Both scans are 1 μm in resolution. Figures 1a and 1c shows a successful cake-in-vial scan without any beam hardening artifact. Porosity and a thin wall of protein solid are both resolved clearly. Figures 1b and 1d show a scan of the same cake sample taken out of the glass vial. Small deformation of the protein solid is captured during the scan as double-edged features, denoted by the red arrows in Figure 1d. The comparison clearly indicates that the scan corresponding to 1b, where the cake is taken out of the glass vial, is undesirable. However, subtle change of the protein solid layers is captured and can be correlated with the mechanisms of cake collapses, due to the high resolution and contrast of XRM.
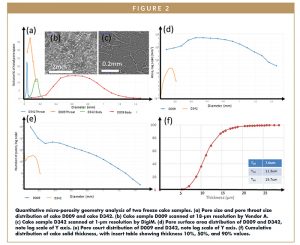
Resolution is very important to how a lyophilized microstructure may be elucidated, as in any imaging study. Figure 2b and 2c show two lyophilized cake samples imaged at 18-μm and 1-μm resolution, respectively. In Figure 2b, a cake sample D009 was imaged at 18-μm resolution. Only the large pores are visible, while a fair amount of microporosity cannot be fully resolved, leaving a lot of voxels (3D version of pixels) with a mixture of cake solid and porosity. In Figure 2c, a cake sample D342 imaged at 1-μm resolution, in comparison, can resolve the thin cake filament with very good contrast and resolution, which is essential for quantitative analysis.
As the first advantage of image-based lyophilized product characterization method, XRM imaging provides direct, non-invasive, explicit and full 3D visualization of lyophilized samples, at resolutions as high as sub-micron.
AI MICROSTRUCTURE QUANTIFICATION
High-resolution 3D imaging, including but not limited to XRM and MicroCT, produces massive volumetric image data with rich microstructure information for lyophilized drug and other solid and semi-solid dosage samples. The management, analysis, and quantification of these data become a non-trivial task. Conventional desktop-based analysis software solutions, commercial or open source, fall short on dealing with these data effectively.
Using a cloud computing interface, DigiM I2S, thousands of 3D volumes can be easily accessed, visualized, searched, and managed via a web-based browser, from anywhere, at any time, using any kind of computing devices.6
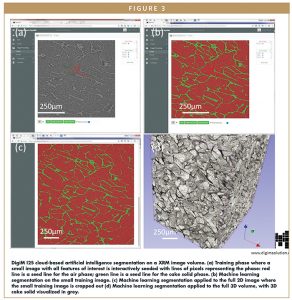
Figure 3a shows a snapshot of DigiM I2S’ AI-based image segmentation interface. The user interactively draws traces of lines as the seed to train the DigiM I2S AI engine. The red trace is the training seed for the porosity, and green trace is the training seed for protein solid. Figure 3b shows the results where the image is classified into two phases, red phase for porosity and green phase for protein solid. If the classification is unsatisfactory, the user can revise his/her seeding strategy by removing, adding, or modifying trace lines.
When the user considers the supervised AI segmentation on the training image is acceptable, Figure 3c, a cloud computing batch session can be launched on the full image volume, through which the 3D cake structure can be reconstructed, as shown in Figure 3d. These 3D reconstructions fully digitize the cake microstructure. Thus, they can be visualized, digitally edited, and quantified.
QUANTIFICATION
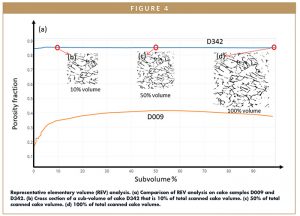
With reconstruction of 3D microporosity and cake structure, quantification is possible. Table 1 summarizes the quantifications computed from the two 3D imaging scans corresponding to Figure 2b and 2c. Resolved porosity of the two cakes are 38% and 84.9%, respectively. Figure 4 shows how porosity varies within each cake sample. Starting with a small sub-volume (eg, 10% of total cake volume image), porosity is computed for each incremental sub-volume. When the porosity reaches a constant volume, i.e., it does not further increase with the increase of sub-volume size, a representative elementary volume (REV) is considered to be obtained. D342 has reached REV with about 30% of total volume, while D009 has not achieved REV even with the full volume. Hence, high resolution XRM imaging compared to MicroCT offers the advantage of more accurate pore structural quantification by capturing more representative REVs.
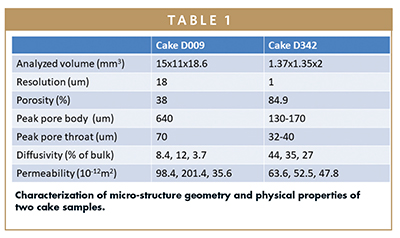
The reconstructed 3D microstructure can be quantified in various ways, such as pore body size distribution and pore throat distribution (Figure 2a), pore surface area distribution (Figure 2d), pore number distribution (Figure 2e), and cake solid thickness distribution (Figure 5f). With the digital microstructure data, a large suite of geometrical characterization can be computed, including but not limited to shape, orientation, aspect ratio, and ferret diameter of pores. While porosity can be obtained with other laboratory measurement methods, with arguably higher uncertainty and more difficulty than imaging analytics methods, pore size distribution, surface area distribution, and cake solid thickness cannot be measured otherwise at all. Full 3D quantification capability is the second distinctive advantage of the image-based lyophilized product characterization workflow.
PHYSICAL PROPERTY PREDICTION
Qualitative visualization at high resolution and quantification in 3D, as discussed in previous sections, are both extremely valuable. However, they are characterizing the structures that are captured at one particular time instance, under fixed physical conditions. By combining the microstructures from the images with computational physics methods, properties under variable physical conditions can be predicted. Variable physical conditions include, but are not limited to, pressure, temperature, fluid flow, and electromagnetic field.
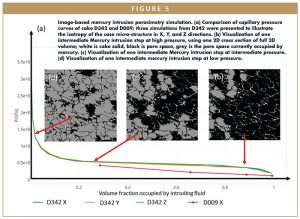
Figure 5 shows a Mercury intrusion capillary porosimetry (MICP) simulation result conducted with DigiM I2S on samples D342 and D009. MICP is based on the premise that a non-wetting liquid (one having a contact angle greater than 90°) will only intrude porous capillaries under pressure. The relationship between the pressure and capillary diameter is described by Washburn as: P=-2*gamma*cos(theta)/r, where P is pressure, gamma is surface tension of the liquid, theta is contact angle between the liquid droplet surface with the solid wall, and r is the radius of the pore.7
As a non-wetting liquid to most materials, mercury must be forced using pressure into the pores of a material. The pore size distribution controlled by pore throats is determined from the volume intruded at each pressure increment. Total porosity is determined from the total volume intruded. The MICP technique is used widely in the industry because of its ease and simplicity. However, for most pharmaceutical materials, it is very difficult to keep sample integrity when it is subjected to high pressure. The sample can deform under moderate-to-high pressure, thus changing the geometry being measured. The sample can also fracture, leading to measurement inaccuracy and failure. These challenges are more pronounced on lyophilized drug samples. An image-based MICP simulation method adapted to the cloud computing environment DigiM I2S circumvented all of the aforementioned problems and avoided the use and disposal of Mercury entirely.8
Figure 5a shows four MICP simulations performed by DigiM I2S. The three curves correspond to the three directions of D342 sample. The three curves have nearly identical shape, indicating isotropic microporosity structure of D342. The result corresponding to the X direction of D009 is also shown. Due to the bigger pores resolved with relatively lower resolution in cake sample D009, only four pressure points are captured with this simulation with much lower threshold entry pressure.
Figure 5b, 5c, and 5d show three intermediate saturation states associated with their corresponding pressure in the MICP simulation of X direction for sample D342. One cross-section is shown from the full 3D simulation at each saturation state. In the simulation, mercury is pushed into the cake sample from left to right. The simulation result curves run from right to left with increasing pressure. At low pressure, Figure 5b, only a small portion of the pore space near the inlet is saturated with mercury (grey), while the majority of the pore space is still empty with only air (black). Cake solid is visualized as white voxels. When pressure increases, Figure 5c, more Mercury is pushed into the cake sample, thus mercury saturation is increased and occupies 70% of the total pore space. Near the final stage at high pressure, Figure 5d, more than 95% of the total pore space is occupied.
Image-based simulation can produce not only pore space distribution characterization of lyophilized samples without risking sample integrity, but also direct visualization to allow elucidation of dynamic transport processes. Other simulations, such as Navier-Stokes-based permeability, Second Fick’s law-based molecular diffusivity, Ohm’s law-based electrical conductivity, Fourier’s law-based thermal conductivity, and linear element-based Young’s modulus, can be computed. With the image-based method, physical properties are derived from the exact same microstructures, which is very difficult, if not impossible, to do otherwise.
FURTHER POTENTIAL & LIMITATIONS
Image-based characterization has great potential in a number of lyophilized drug product characterization situations. Cake integrity and purity can be non-invasively monitored. Inter-batch and intra-batch homogeneity can be quantified. Correlations of sample microstructure with lyophilization process can be mechanistically understood, parameterized, and optimized, including sublimation rate, primary and secondary drying optimization via annealing, and controlled ice nucleation. Specifically, if we can derive mass transfer and heat transfer coefficients from the cake structure and couple them with lyophilizaiton modeling tools, the results would be extremely helpful for cycle development and scale up.9 Optimistically, this approach could be used for stability prediction and failed batch diagnostics. Like any characterization effort, a number of limitations need to be kept in mind. A successful image-based characterization project requires an optimal balance between resolution and the number of samples studied to ensure representativeness. Finite resolution limits the smallest feature that can be studied. Furthermore, due to the high price of imaging devices, sophistication in the imaging experiment, and the massive amount of data derived, the cost of image-based analysis is not trivial. However, as the image data progressively develops into a digital drug database, with more insight, faster drug evaluation, and facilitation of new formulation design, the initial imaging cost will be marginalized. Last but not least, the management, visualization, and quantification of simulation of massive amount of data is often overlooked and limits the utility of 3D imaging data. A dedicated software and computing hardware platform is often required, where a cloud computing solution, such as DigiM I2S, offered both as software as a service (SaaS) and on-premise installation, has a great advantage.
REFERENCES
- Wang W. Lyophilization and development of solid protein pharmaceuticals. Int J Pharm. 2000;203(1):1-60.
- Pikal MJ. Mechanisms of protein stabilization during freeze-drying storage: the relative importance of thermodynamic stabilization and glassy state relaxation dynamics. In: Rey L, May JC, eds. Freeze-Drying/Lyophilization of Pharmaceutical & Biological Products. 3rd ed. New York Informa Healthcare;2010.
- Pisano, R., Barresi, A., Capozzi, L., Novajra, G., Oddone I & Vitale-Brovarone, C. Characterization of the mass transfer of lyophilized products based on X-ray micro-computed tomography images, Drying Technology, 2017; 35:8, 933-938.
- Haeuser C, Goldbach P., Huwyler J, Friess W, Allmendinger A. Imaging Techniques to Charactorize Cake Appearance of Freeze-Dried Products. Journal of Pharmaceutical Sciences. 2018;107:2810-2822.
- X-ray microscope Wikepidia page. https://en.wikipedia.org/wiki/X-ray_microscope. Accessed 2/16/2019.
- Zhang, S., Byrnes, A. P., Jankovic, J. & Neilly, J. Management, Analysis and Simulation of Micrographs with Cloud Computing. Microscopy Today. March 2019; 27:2:26-33.
- Washburn, E.W. Note on a method of determining the distribution of pore sizes in a porous material: Proceedings of the National Academy of Science, 1921;7:4:115-116.
- Hilpert, M., & Miller, C.T. Pore-morphology-based simulation of drainage in totally wetting porous media. Advances in Water Resources. 2001;24:3-4:243-255.
- Chen, X., Sadineni, V., Maity, Mr., Quan, Y., Enterline, M., and Mantri, R. Finite Element Method (FEM) Modeling of Freeze-drying: Monitoring Pharmaceutical Product Robustness During Lyophilization AAPS PharmSciTech, Vol. 16, No. 6, December 2015;16:6:1317-1326.

Dr. Shawn Zhang is Founder and Managing Partner of DigiM, where he leads DigiM to becoming the platform technology pioneer in microstructure physics, characterization, and design with applications. He earned his PhD in Computational Physics from Rutgers University.

Dr. John Goldman is a Process Development Scientist at Moderna Therapeutics, leading its mRNA lipid nanoparticle drug product lyophilization development. Dr. Goldman earned his PhD in Chemical Engineering from Carnegie Mellon University.

Dr. Xiaodong Chen is a Senior Research Investigator at BMS, responsible for developing, recommending, and implementing development strategies of biologics drug product formulation, device and primary packaging, tech transfer to commercial manufacturing, and registrational filing. He earned his PhD from the Ohio State University.

Dr. Jasmine Rowe earned her PhD in Chemical Engineering from the University of Texas at Austin. She leads an Applied Engineering group within drug product development at BMS, where her group focuses on the development and integration of engineering tools and models for both oral solids and parenteral products.

Sam Lin is a Software Development Manager with DigiM. As one of the chief architects of the first cloud-based image processing platform, he leads the design and development of DigiM I2S. He also champions AI-based image processing platform technology since he joined DigiM in 2015. He earned a Computer Science degree from QingDao University.

Dr. Liping Zhou joined DigiM Solution with 15+ years of experience in the pharmaceutical and biotech industry, including Novartis and Ipsen. She earned her PhD in Chemistry from the University of Connecticut and has nearly 20 publications in peer-reviewed journals and book chapters to her credit.
Total Page Views: 6642