Issue:November/December 2018
HUMAN MICROBIOME - Advancing New Frontiers in a Rapidly Emerging Market
INTRODUCTION
The field of Human Microbiome research and development is apparently one of the most popular hubs of the biotechnology industry. While the Human Microbiome Project, MetaHIT and other huge studies of human microbiota, have garnered a lot of attention over that past few years, the microbiome space has literally exploded in terms of both basic and applied biomedical research.
This report focuses on biomedical aspects of research, development, and commercial endeavors in the human microbiome space. It includes essential background information, evolution of the field, advances in basic research, events in the emerging commercial market, deal activity, interviews with experts, and trends in microbiome research and commerce. Primary sources of information for this report include the scientific literature, discussions with experts, and an online survey of individuals working in this space.
In 2009, a PubMed search on the term human microbiome yielded 579 citations, with a radical increase to 4,490 by 2014. Not only has the field seen massive investments from Venture Capital firms and Angel Investors, showing keen interest, but there has also been a flurry of deals and collaborations with the expected flux and knowledge exchange. Another milestone achieved was the first major microbiome IPO issued.
HISTORY & EVOLUTION
After pioneering several milestones in the field of genomics, Craig Venter led an expedition to collect samples of marine bacteria and sequence them en masse using Sanger shotgun sequencing technology. Venter and his team identified more than 1,800 species, a groundbreaking feat that established metagenomics as a sustainable field for investigation and commercial involvement. The arrival of next-generation sequencing (NGS) technologies with the ensuing emphasis on short hypervariable regions of microbial 16S rRNAs enabled the Human Microbiome Project and other similar efforts to comprehensively characterize the human microbiome to encourage full participation in the field. Initial efforts in microbiome research and development helped to identify several enterotypes (classification of living organisms based on its bacteriological ecosystem in the gut microbiome); a solid foundation for subsequent work in personalized microbial medicine. Evidence shows that individual enterotypes seem to change in response to dietary alterations. Another observation is that a well-balanced microbiome can help to maintain weight, whereas disruption or dysbioses can lead to obesity. Several other correlations have been made between dysbiosis and disease. The current wave of increased commercial activity was born out of several studies showing that patients suffering from recalcitrant Clostridium difficile associated diarrhea could be treated with a fecal transplant from a “healthy” matched donor. Antibiotic treatment temporarily alters the gut microbiome and allows the normally dormant commensal C. difficile to overgrow. This fecal microbiota restoration therapy helps to restore balance with success rates of up to 100%.
Advances in Research on the Human Microbiome Initially NGS sequencing of metagenomes was based on the Roche 454 pyrosequencing platform. 454 Life Sciences will no longer support the platform after 2016. The need for more time-saving and cost-effective alternatives shifted attention to mainly Illumina’s faster and cheaper short-fragment sequencing systems. Some researchers favor working with combinations of sequencing platforms depending on the application. Other researchers prefer Pacific Biosciences’ platform; the latest release P6-C4 provides impressively long reads averaging 10,000 to 15,000 bases. Data analysis remains a challenge in the microbiome space. Informatics workflows can be either gene-centric, preferred when addressing high complexity applications; or assembly-based, favored for lower diversity applications. Either choice requires further selections downstream from the branch point.
Quite a few substantial advances in data analysis have been made recently yet inconsistency among results of metagenomics analyses from different laboratories and technologies/platforms is a huge challenge. Protocols must be standardized and results made consistent across laboratories and technology platforms. In a collaborative effort, researchers from Human Longevity, Inc. (HLI) and the J. Craig Venter Institute (JCVI) recently published a paper calling for standardization of practices across laboratories. The paper highlighted inconsistencies in microbiome work from different library preparation methods and data analysis.
COMMERCIAL ACTIVITY
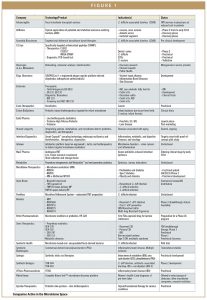
This report profiles the activities of 28 microbiome companies, most of which are engaged in developing therapeutics for various diseases. The most frequent target indication is C. difficile gastroenteritis; not surprisingly the gut microbiome is the research focus of the majority of the companies. Other diseases include inflammatory
bowel disease, irritable bowel syndrome, acne, and diabetes. Particular programs are directed at larger therapeutic areas, such as neurodevelopmental disorders, autoimmune disease, metabolic disorders, and infectious disease. One company works to develop synthetic oral biotics to address the challenges of inborn errors of metabolism. Several companies are committed to developing more refined alternatives to FMT, including oral preparations and storage devices.
Diagnostics in development range from biomarkers to monitor mucosal healing in IBD, to oral preparations for sampling the gut. The report also describes 15 recent microbiome-related deals, including four research partnerships between small companies and Big Pharma. These include an unidentified global Pharma company and Janssen Research and Development unit. The APC Microbiome Institute University of College Cork, Ireland, is collaborating to support two small companies. The National Institute of Allergy and Infectious Diseases (NIAID) and Swiss Foods Company Nestlé are providing collaborative support to smaller companies.
In an online survey of 119 individuals active in the microbiome space, more than half of respondents work in academia compared to less than half who work in commerce. Nearly one-third are addressed as research/development manager, group leader or supervisor. Almost half stated their work involved analytical methods to detect or describe microbiomes in individuals or populations. About two-thirds work on the gut microbiome and by therapeutic area, another two-thirds focus on inflammation. The majority of respondents work on cancer.
None of the participants expect a change in the research focus of their company, and two-thirds expect an increase in the company’s microbiome efforts over the next 2 years. One-third of respondents opined that sufficient microbiome-related information has been obtained to warrant translational efforts, while less than one-third disagreed. Nearly one-third use microarrays to detect dysbiosis, while less than a third use short-read next-generation sequencing. Nearly half of respondents agree that the next decade will see an avalanche of new personalized biotherapeutics.
TRENDS & CONCLUSIONS
Our interviews and survey results show that persons working in the microbiome field are highly optimistic about the relevance of their work to the future success of microbiome research and development. The market is growing tremendously, and microbiome market potential is projected to rise from $294 million in 2019, to $658 million by 2023. Several research efforts are geared toward establishing cause over correlation regarding dysbiosis-related disorders. Groups and consortia have called for a unified global microbiome effort possibly to promote consistency of results and standardized protocols. This is requisite to maintain the highest standards of quality control for human health.
The term human microbiota refers to the 10 to 100 trillion symbiotic microbial cells harbored by an individual, mainly bacteria in the gut; the human microbiome consists of the genes these cells harbor. An unfavorable change of the gut microbiota composition is called dysbiosis, which leads to an overgrowth of potentially pathogenic bacteria (pathobionts) and a decrease in the number of beneficial bacteria (symbionts). Preceding the current wave of interest and investment in microbiome research and development, early work in the field was hampered by an inability to culture the majority of microbes found in humans. Craig Venter, whose application of shotgun DNA sequencing contributed immensely to the Human Genome Project, also played a key role in triggering the current microbiome gold rush. In 2003, he pioneered the Global Ocean Sample Expedition and led a group that traveled the seas collecting samples of bacteria, later identified by Sanger shotgun sequencing. In 2004, Venter’s group published results from a pilot study on samples taken from the Sargasso Sea near Bermuda. They identified about 1,800 species, which included 148 new lineages of microbials. That same year, Jo Handelsman in a seminal paper defined Metagenomics as “the genomic analysis of microorganisms by direct extraction and cloning of DNA from an assemblage of microorganisms,” thus, introducing a method of analysis that allowed the application of genomics to uncultured organisms. Metagenomics (also referred to as environmental and community genomics) is another important term to use concerning the microbiome. Simply put, this means that a mixture of DNA from different sources, microbes, in this case, could be cut in pieces, the fragments sequenced, and results deciphered to provide information on the types of organisms contributing to the mix. 454 Life Sciences (a Roche company) brought the first next-generation sequencing technologies to market, with the overall approach being introduced in 2005. A major milestone in the field was the application of 454 Life Sciences’ next-generation Pyrosequencing technology to the analysis of bacteria in a deep mine ecosystem by Penn State University scientists. Another major boost to the field was when Illumina and others developed alternative methods and instruments more suitable to gathering large amounts of sequence data at reduced costs in a shorter time frame. From classical Sanger sequencing, the field progressed into determining sequences of entire 16S rRNA genes, which in turn evolved into the current more efficient sequencing of small hypervariable regions of these genes.
This executive summary is based on the following market research report published by Insight Pharma Reports: The Human Microbiome: Advancing New Frontiers in a Rapidly Emerging Market by Flo Orim, MD. For more information, visit www.insightpharmareports.com.
To view this issue and all back issues online, please visit www.drug-dev.com.

Dr. Flo Orim is an experienced physician and cancer researcher who brings a passion for the simple communication of complex science to her work as a medical writer. With an MBBS from University of Jos, Nigeria (1999) followed by years of clinical practice, she made a brief foray into managed care/health insurance and rediscovered her love for communication as a lecturer at the medical school in University of Abuja, Nigeria. She was awarded a Japanese Government scholarship and earned her PhD in Medical Sciences (Radiation Research and Life Sciences) from The Atomic Bomb Disease Institute, Nagasaki University (2013), delving into medical/academic writing during the course of graduate school. She then completed postdoctoral training in Molecular Pathology at Aichi Cancer Center Japan (2016). Dr. Orim has several publications, gaining academic and medical writing experience across the clinical and basic medical sciences as well as the life sciences and biotechnology. Dr. Orim currently works as a medical writer and editor in Nagoya, Central Japan.
Total Page Views: 5643