Issue:September 2020
GENE-EDITING TECHNOLOGY - The Importance of Assessing Off-Target Effects of CRISPR Gene Editing
INTRODUCTION
“It feels like my bones are being sawed with a rusty blade, for hours” says Alice (not real patient’s name) – a woman in her 30s who has lived with a lifelong condition, called sickle cell disease (SCD). “I just have to tough it out. I try warm soaks, heating pads, and deep breathing to soothe the agony. It’s hard to get the right care and pain relief. You go into the emergency room and you don’t know if you’re going to leave alive. Every time, it’s a battle” she says.1 Pain is the hallmark of SCD, and is caused by deformed, sickle-shaped red blood cells clumping together and blocking capillaries. This results in severe pain and swelling in affected body parts. SCD can be life-threatening. In fact, half of the patients that make it to adulthood are dead by their early 40s.

THE DISCOVERY OF CRISPR
In the early 1990s, a scientist completing his PhD in Spain discovered genetic sequences in archaea that were repeated up to 600 times in a row. This corroborated a similar discovery in bacteria by Japanese scientists in the late 1980s. These repeats were later found to be part of a prokaryotic immune system. They are used to store genetic information on viruses to which the organism and its progenitors have been exposed, and prime the organism to defend itself against these threats in the future.2 At the time, few people would have imagined that this finding, which later became known as CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats), would lead to further developments that would become the dominant gene editing technology and have a major impact on the medical sciences.
CRISPR gene editing is a technology that is being used by scientists to make precise, permanent changes to DNA in animals and plants. CRISPR can be used to make these edits at a single, specific location. It is currently being evaluated in early phase clinical trials for several disorders caused by a single gene mutation, including SCD and beta-thalassemia. As the technology evolves, we will likely see more applications beyond treating these types of disorders, as well as use of the technology in agricultural science to improve crop yields and resilience and boost nutritional value.
WHAT IS CRISPR GENE EDITING?
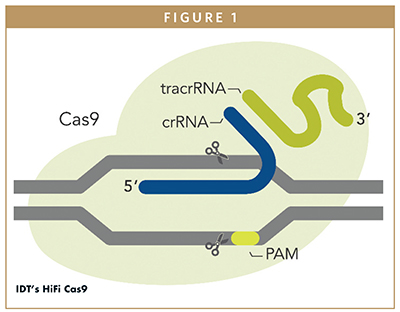
CRISPR gene editing is based on a natural defense mechanism, used by bacteria and archaea. It is composed of two components: a nuclease (eg, Cas9), responsible for the cleavage of double-stranded DNA, and an RNA guide, which provides specificity by aligning the nuclease–RNA complex to the desired target DNA. The technology is currently generating a lot of excitement because it is more cost effective, rapid, and easy to use than previous gene-editing techniques, including zinc-finger nucleases and TALENs. As the most commonly used enzyme in CRISPR gene editing, Cas9 is often applied to target and edit precise stretches of DNA and has been a key focus of our work at Integrated DNA Technologies (IDT).
THE CAS9 ENZYME
The first case of a nuclease engineered for the CRISPR technique was the Cas9 system from Streptococcus pyogenes (S.p.), which was described in 2012.3 This CRISPR system, composed of wild-type (WT) Cas9, and two guide RNA molecules fused into a single guide RNA (sgRNA), has been successfully used for gene-editing purposes in mammalian cells.
CRISPR/Cas9 gene editing offers unparalleled genome editing efficiency and can function in various cell types and species. However, despite the improved specificity of CRISPR in recognizing target sequences when compared with previous gene-editing techniques, DNA cleavage may still occur at unintended sites that are similar but not identical to the desired target site. These are known as off-target effects (OTEs) and may account for more than 50% of the editing delivered by WT Cas9.4 Although these events mostly occur in areas of the genome believed to have no function, there is always the risk that they may lead to unintended, adverse consequences. Many researchers have tried modifying the guide RNA and created improved-specificity mutant Cas9 proteins to reduce OTEs, but these alterations also usually reduce on-target editing performance.
ENGINEERING A HIGH-FIDELITY CAS9 ENZYME
Our work at IDT has focused on developing and improving the tools available for biomedical scientists to conduct their research in the field of gene editing. A recent breakthrough was the development of a new high-fidelity Cas9 enzyme, known as HiFi Cas9. We achieved this by devising an unbiased bacterial screen to isolate ribonuclease protein (RNP)-delivered Cas9 variants that displayed highly specific cleavage with minimal OTEs, while retaining full on-target nuclease activity in line with that of WT Cas9. The results of this work and clinical applications were published in Nature Medicine in 2018.5
As the most active and specific high-fidelity Cas9 variant available when delivered as an RNP complex, it is ideal for use in clinical studies. This led IDT to partner with Aldevron to supply a Good Manufacturing Practice (GMP) grade of the enzyme for clinical use.
Excitingly, Dr. Matthew Porteus and his team from Stanford Medical School have been using this high-fidelity enzyme successfully in their preclinical studies, ahead of a proposed Phase I SCD clinical trial. The trial is investigating the potential for a gene-editing therapy to correct the SCD-causing point mutation in the human beta-globin gene (HBB). The HiFi Cas9 enzyme was used to show that the sickle HBB allele could be efficiently corrected, while simultaneously reducing problematic off-target editing from ~30% to less than 1%, in the preclinical studies (compared with using WT Cas9). The Aldevron GMP HiFi Cas9 enzyme provides similar impressive results and will ultimately be used in the clinical trial for SCD, as well as other monogenic hematologic disorders.
“We performed an unbiased evaluation of several versions of high-fidelity Cas9 enzymes in primary human stem cells. We have been very impressed with the characteristics of this new IDT enzyme. Unlike other versions, this version consistently gives us high on-target editing activity, while having low off-target activity. Because of the retained, excellent on-target activity and improved specificity profile, we are excited to use this version in our future experiments focused on developing novel genome editing-based therapies for several diseases with unmet medical needs,” said Dr. Porteus.
A CAUSE FOR CONCERN
While CRISPR gene editing is being considered for numerous applications in animals and plants alike, and clinical trials have already begun to recruit participants and administered investigational therapies, one cause for concern remains the potential for OTEs. Such concerns in this field are not new. For viral vector-based gene editing, researchers were worried about the potential for a mutagenic vector to integrate into the genome. For zinc-finger nucleases, there was concern that the method could cause OTEs according to cell-based experiments.6 For CRISPR, research continues to suggest that OTEs require further analysis and review. The nucleases used in CRISPR gene editing cut the DNA and create double stranded breaks, and the sgRNA directs the enzyme complex to the correct location in the genome. However, these guides may also cause breaks to occur at unintended sites. While these may not have a noticeable impact and may not even be detected, it remains important to understand the frequency and nature of these effects and more accurately predict occurrence to minimize risk when using this technology for medical purposes. To that point, there are numerous solutions being investigated by scientists. In early 2018, the US National Institutes of Health (NIH) announced the launch of the Somatic Cell Genome Editing (SCGE) program. The program will run for 6 years and will support researchers bringing gene- editing therapies to patients and developing novel assays for assessing OTEs.
WHERE WE ARE TODAY?
In addition to improving enzyme specificity, researchers have been using computational algorithms to predict which regions of the genome might be at risk for OTEs. These algorithms usually identify OTEs but often provide either too many sites to study or are filtered too much and so miss important sites. In response to these uncertainties, IDT launched a new product called rhAmpSeqTM that can be used in preclinical risk assessments. A number of pharmaceutical and biotech companies as well as academic centers doing translational medicine studies are already using rhAmpSeq to help reduce the work-load of monitoring OTEs while optimizing the performance of their CRISPR therapeutic protocols. The technology enables a researcher to rapidly characterize hundreds to thousands of individual off-target and on-target sites from a single DNA sample using modern multiplex next-generation sequencing (NGS) methods.
Even with tools like rhAmpSeq, we are still in the early years of CRISPR technology, and there is still much to learn. As the technology is optimized to minimize OTEs, there will still be a need to acknowledge that a certain degree of risk is inherent in many promising potential and proven successful treatments and that CRISPR is no different.
HOW MUCH RISK ARE WE WILLING TO ACCEPT?
With the first human trials using CRISPR to treat several diseases already underway, a key question remains: what does greater awareness of OTEs mean for clinical applications of CRISPR and how might it improve safety profiles for investigational and approved therapies?
It is not uncommon for new medical therapies to be approved for use despite their potential to cause harm. This is because of the benefit/risk ratio, whereby benefits of the therapy in addressing the burdens and harms of disease – including the likelihood of death, are weighed against the potential risks and actual burdens and harms of the treatment. Should the benefits exceed the risk, the therapy is deemed appropriate and acceptable. An example of such an approved treatment is the chemotherapy drug, cisplatin. Although a known mutagen, cisplatin is used to treat several ailments, including life-threatening cancers.
A challenge for CRISPR will be that adverse effects may not become known for many years, and risks may vary between individuals because all of us have our own unique genome (different DNA = different risk profile). Despite these and other challenges, the potential of CRISPR gene editing is too great to ignore. It offers people suffering from debilitating and uncurable diseases the hope that one day there may be a one-time treatment that proves to be a lifetime cure. It also offers these same individuals the hope that their disease may one day have a treatment plan unique to them and their genetic code, realizing the dream of truly personalized medicine. In addition to applications in human medicine, CRISPR gene editing has implications and potential uses for agriculture and food security, which could be particularly important considering the challenges expected with projected population growth and climate change. However, with such great hope and promise, comes challenges both in the laboratory and beyond.
OVERCOMING THE CHALLENGES OUTSIDE OF THE LAB
CRISPR is currently being evaluated by researchers, clinicians, and ethicists to assess the potential repercussions of the technology. It will be important that these discussions involve laypersons so that public opinion of the technology is based on informed facts and not uninformed rumors. A paper issued in Frontiers in Public Health in 2017 published results from an online survey of 2,493 respondents and found that while the mention of explicit risks dampened enthusiasm for the use of CRISPR technology, respondents remained largely supportive of the research.7 Interestingly, the researchers noted that the use of common metaphors (editing, engineering, hacking, modification, and surgery) used to describe genetic modification did not influence attitudes. However, these findings only represent a snapshot of attitudes and it is important to remember that the discussion and information available will evolve over time. A key question remains: will the discussion and information available keep up with the pace of scientific discovery?
Publicly available scientific information can be difficult to navigate, sometimes inaccurate and oftentimes complicated to understand for the non-expert reader. We have seen before, in the case of genetically modified organisms (GMOs) for example, that an uninformed or misinformed public can have negative implications for scientific progress. It is therefore paramount that the societal debate keeps up with the rapid pace of research and that the public understands both the benefits and risks of the technology and is confident that it will be used to reduce the global burden of disease and suffering associated with disorders like SCD, cystic fibrosis, and Huntington’s disease. Like other potentially powerful technologies and scientific breakthroughs, gene editing raises legitimate questions and fears about possible risks and misuse. Given the relative technical ease and low cost of CRISPR gene editing, this technology is likely to find widespread use in both medicine and agriculture. To alleviate fears and benefit from the opportunities afforded, open, transparent and continuous debate is required, as well as education of stakeholders – from scientists and governments to civil society and local communities. Without these discussions, and additional efforts to ensure consumer education and engagement, we run the risk of failing to unlock CRISPR’s true potential. As leaders in the provision of CRISPR technologies, we strongly promote and support efforts to raise awareness, education, and discussion regarding CRISPR technology among both multidisciplinary experts and the general public.
REFERENCES
- Begley S. ‘Every time it’s a battle’: In excruciating pain, sickle cell patients are shunted aside. STAT. 2017.https://www.statnews.com/2017/09/18/sickle-cell-pain-treatment/(accessed November 2019).
- Ishino Y, Krupovic M, & Forterre P. History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology. Journal of Bacteriology. 2018; 200: e00580-17.
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337: 816–821.
- Zhang, X.H., Tee, L.Y., Wang, X.G., Huang, Q.S., & Yang, S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic. Acids. 2015;17: 4.
- Vakulskas CA, Dever DP, Rettig GR, Turk R, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. 2018;24: 1216–24.
- Keep off-target effects in focus volume. Nature Medicine. 2018;24:1081.
- Weisberg SM, Badgio D and Anjan C. A CRISPR New World: Attitudes in the Public toward Innovations in Human Genetic Modification. Front Public Health. 2017;5:117.

Mark Behlke, MD, PhD, is Chief Scientific Officer at Integrated DNA Technologies (IDT) and is an internationally recognized expert in the field of nucleic acid technologies and an inventor cited on more than 50 US Patents. In nearly 25 years at IDT, Mark has overseen research programs and new product development in the areas of DNA thermodynamics, gene synthesis, probe chemistry, qPCR, NGS, antisense, RNA interference, and CRISPR genome editing. Mark joined IDT as an R&D consultant in 1995 and was named VP, R&D in 1996. He was promoted to CSO in 2013.
Total Page Views: 4718