Issue:June 2021
GENE-EDITING TECHNOLOGY - Expanding the CRISPR Toolbox for Genome Editing
INTRODUCTION
Genome editing technologies have existed since the 1990s but have recently made a surge, largely due to the discovery of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology in 2012.1 CRISPR genome editing is being investigated by researchers to make precise and permanent changes to DNA in animals and plants. Current applications include efforts to increase crop resilience and yield, to help contribute to feeding the growing world population. With two billion more people to feed by 2050, solutions are needed to increase food production without overwhelming the planet and genome editing is one practical option. In addition, it offers patients suffering from untreatable diseases hope that one day there will be a one-time cure available to them.
AN OVERVIEW OF THE TECHNOLOGY
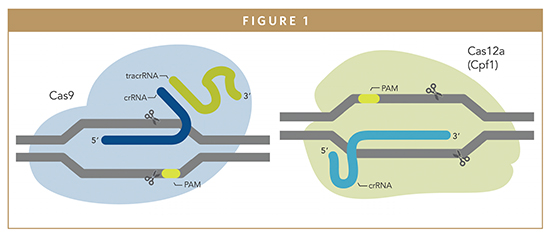
CRISPR technology comprises two components: a nuclease, e.g., Cas9, which acts like a pair of scissors and is responsible for cleavage of double-stranded DNA, and a single guide RNA (sgRNA), which forms a complex with the nuclease and guides it to the target site. The technology is groundbreaking because it can be designed to make a break at a specific target sequence in the DNA within a living cell, allowing researchers to modify practically almost any locus in the genome of any organism.2 It is also more cost effective, precise and faster than previously available editing methods like zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs).
As the most commonly used nuclease in CRISPR gene editing, Cas9 can be applied to target specific stretches of genetic code and edit DNA at precise locations, which may make it possible to correct the DNA mutations underlying diseases like sickle cell disease (SCD), Huntington’s disease and cystic fibrosis. However, DNA cleavage and editing may occur at additional sites in the genome that have a similar DNA sequence from that of the intended site. Such events are called “off-target effects” (OTEs). Use of wild type (WT) Cas9 to target certain gene locations has been shown to lead to more than 50 percent of the edits being unspecific.
While CRISPR/Cas9 gene editing offers unparalleled genome editing efficiency, and can function in various cell types and species, challenges remain due to low target site specificity and on-target efficiency. Many researchers have tried modifying the guide RNA and mutant Cas9 proteins to improve target specificity, but these alterations often also reduce on-target editing performance.
REFINING THE CAS9 ENZYME AND INTRODUCING CAS12A
At Integrated DNA Technologies (IDT), we have successfully engineered a high-fidelity Cas9 that has greater targeting specificity than the WT Cas9, while retaining nuclease activity that is comparable with WT Cas9. In fact, our HiFi Cas9 is the most active and specific high-fidelity Cas9 enzyme available, when delivered as an RNP complex, which provides optimal targeting specificity.
Although Cas9 is the most commonly used CRISPR nuclease, others do exist. Cas12a (formerly known as Cpf1) has recently emerged as an alternative to Cas9 in the toolbox of CRISPR-based genome editing tools.3 There are several unique features of Cas12a that distinguish it from Cas9; most notable is the fact that Cas12a targets AT-rich regions of the genome, compared with Cas9, which targets GC sequences. However, wild type Cas12a suffers from lower nuclease activity than Cas9.
At IDT, we recently developed the Alt-R CRISPR-Cas12a (Cpf1) Ultra system. The new variant, Cas12a Ultra, has enhanced editing activity and can be used across a broader temperature range than the WT Cas12a enzyme, making it useful for genome editing in a variety of organisms, including plants and mammals. Given the unique characteristics demonstrated, Cas12a has several potential uses for gene editing, which differ from Cas9.
APPLICATIONS FOR CAS12A & BEYOND
In AgBio applications, gene editing using Cas12a could be used to make crops more resilient, increase yields and boost nutritional value. Mushrooms with longer shelf lives, potatoes low in the possible carcinogen acrylamide, and soy beans that produce healthier oil are already being developed and tested, to name just a few.4 The technology can also be applied to livestock and research is underway to better understand how gene editing might be used to help farmers in Africa breed more productive chickens and cows.5

Interestingly, research is being conducted to analyze the impact of using gene editing to prevent malaria-type disease transmission. Although current efforts and tools to prevent transmission must remain in effect, full eradication will require technological advances. And gene editing is a tool being investigated. One way to do so that has been demonstrated to work is targeting female mosquitoes that are able to transmit malaria and using gene drives – making inheritable edits to their genes – to render them sterile or skew them towards mostly producing male offspring, unable to transmit the disease. However, eradicating a disease is one thing, practically eradicating a vector species (e.g. through an all-male population) is another. Along with the widespread potential impacts and other unknowns of these efforts, ethical, legal, social and regulatory implications need to be carefully considered.
These considerations also need to encompass the rapidly growing list of new Cas enzymes being developed, other than Cas9 and Cas12a, and the applications that are being developed for them. These other enzymes can work alongside Cas9 or serve different functions. For example, Cas13 is a unique enzyme in that it targets RNA not DNA. Once it is activated by a single strand RNA (ssRNA) sequence that matches its CRISPR RNA (crRNA) spacer, it unleashes a non-specific ribonuclease activity and destroys all RNA regardless of sequence. This characteristic has been harnessed in vitro for precision diagnostics.6 Mammoth Biosciences are developing a CRISPR diagnostic platform, which may include Cas12a, Cas13a, and Cas14. The use of multiple Cas nucleases means that a larger range of sequences can be recognized, thus allowing the panel to provide diagnostic information on a broader range of diseases, conditions, and pathogens. The Cas13 crRNA spacers can be designed to DNA orthologs and generated through a transcription reaction in vitro. However, increased CRISPR performance has been observed with RNA synthesized by providers such as IDT.7
WHERE DO WE GO FROM HERE?
While there are many ever-improving tools available to scientists performing ground-breaking research, and the potential of CRISPR genome editing appears limitless, there remain challenges that need to be overcome to realize the technology’s full potential. Most importantly is the off-target editing that can occur. However, there are other technical questions that remain less known. These include the potential that more than half of humans may have pre-existing humoral and cell-mediated adaptive immune responses to CRISPR/Cas9 proteins, which are mostly derived from S. aureus and S. pyogenes – known bacteria that cause infectious disease in humans at high frequency. Another challenge to overcome relates to the delivery of these tools into cells and organisms. With time and continued research, these challenges could be overcome, opening up the possibility to realize the technology’s full potential.
REFERENCES
- Carroll D and Alta Charo R. The societal opportunities and challenges of genome editing. Genome Biology. 2015; 16: 242.
- Carroll D and Alta Charo R. The societal opportunities and challenges of genome editing. Genome Biology. 2015; 16: 242.
- Swarts DC and Jinek M. Wiley Interdiscip Rev RNA. 2018; 9(5).
- Gene Editing for Good. Foreign Affairs. Available at: https://www.foreignaffairs.com/articles/2018-04-10/gene-editing-good. Accessed August 7, 2019.
- Gene Editing for Good. Foreign Affairs. Available at: https://www.foreignaffairs.com/articles/2018-04-10/gene-editing-good. Accessed August 7, 2019.
- Pawluk A. CRISPR Systems: What’s the Difference? Cell. https://www.cell.com/pb-assets/products/research-arc/infographics/CrisprVizInfo_vol1a.pdf. Accessed February 2020.
- Abudayyeh O and Gootenbrg J. Zhang Lab. Tips and Tricks for Cas13. Available at: https://zlab.bio/cas13 Accessed February 12, 2020.

Ashley Jacobi is a Senior Staff Scientist in the Molecular Genetics Research and Development group at Integrated DNA Technologies (IDT). Ashley has been with IDT for 14 years, and during that time, she has been an author on 18 manuscripts published in peer-reviewed journals, contributed to numerous patent applications, and has presented at a wide variety of international biomedical research conferences. She has been conducting research in RNAi and antisense oligo technologies, but more recently has been focusing on the development of novel CRISPR RNA sequences and modification patterns that allow for more efficient and specific cleavage by the S.p. Cas9 and A.s. Cas12a CRISPR nucleases.
Total Page Views: 3644