Issue:November/December 2022
DRUG DISCOVERY - Leveraging the Human Gut as a Drug Discovery Tool
INTRODUCTION
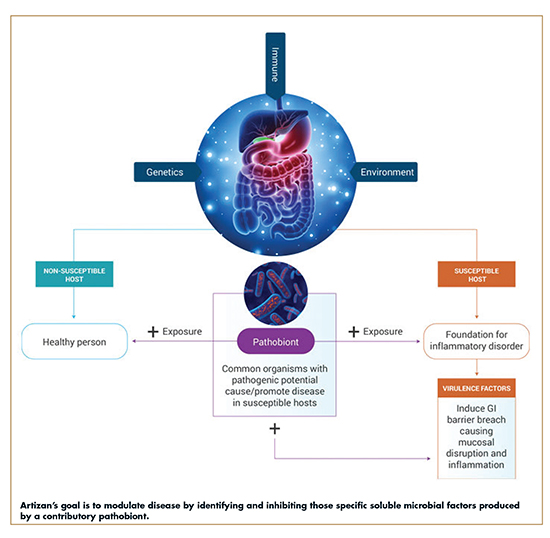
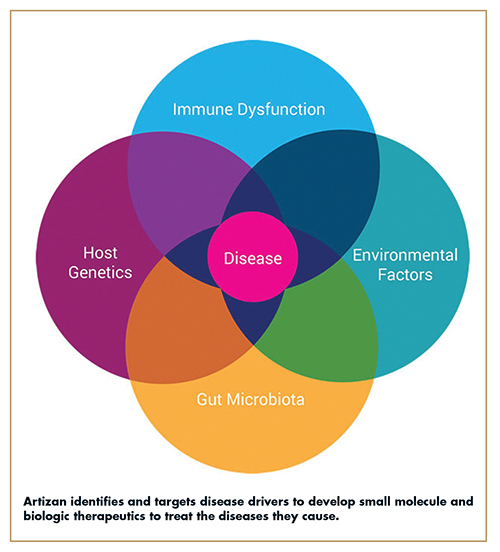
A confluence of factors from host genetics, immune dysregulation, environmental triggers, and gut dysbiosis all contribute to a disease developing in an individual. Given certain genetics and environmental factors, some people are susceptible to organisms (or “pathobionts”) in the gut that can cause disease, while others are not. Pathobionts are defined as bacteria present in the “normal” human intestine that have the potential to cause or drive disease development, and therefore share features with both commensal symbionts and pathogens.1 For example, Segmented Filamentous Bacteria (SFB) are common members of the mouse microbiota that exacerbate the development of autoimmunity, and Helicobacter species drive the development of colitis in genetically susceptible mice.2 SFB and Helicobacter species therefore represent classical pathobionts. In patients susceptible to disease, exposure to pathobionts induces intestinal dysbiosis and triggers an inflammatory cascade that leads to the development and progression of diverse chronic diseases and certain cancers.
Inflammatory bowel disease (IBD) is one example of a disease driven by such factors for which currently available treatments are not a viable solution for all patients. While there have been tremendous advances in IBD therapies, approximately 30% of patients are unresponsive to initial biologics therapy and, even among responsive patients, up to 10% stop responding to treatment annually.
With an initial focus on IBD, that is, Ulcerative Colitis and Crohn’s Disease, Artizan Biosciences has an exclusive license to the patented technology, IgA-SEQ™, invented by its founders to identify putative disease-driving bacteria through the examination of fecal samples from patients with IBD. By leveraging this techonology, Artizan uses the human gut as a drug discovery tool. This process has led to a new class of transformative precision therapeutics that disrupt inflammatory disease triggers – gut exclusive microbial metalloprotease inhibitors (GEMMI).
THE MICROBIOME & HOST INTERATIONS
The composition of the intestinal microbiota varies substantially between individuals and has significant effects on host physiology and disease susceptibility.3-7 A major mechanism by which the microbiota impacts the host is through its interactions with the intestinal immune system. These organisms modulate the intestinal immune system and affect disease risk through chronic stimulation of specific immune responses, of which can be both beneficial and detrimental to the host. In mice, for example, Clostridia species induce the expansion of regulatory T cells and suppress allergic responses and intestinal inflammation, Segmented Filamentous Bacteria (SFB) induce T helper 17 responses, exacerbate the development of arthritis, and protect against the development of diabetes.8-11 Alterations in the composition of the microbiota, sometimes referred to as “dysbiosis,” are known to drive development of both inflammatory and non-inflammatory diseases, including IBD, metabolic diseases, and autoimmunity.12 IBD is a chronic, relapsing chronic inflammatory disease for which there is no cure. In IBD, it is believed a dysbiotic intestinal microbiota plays a key role in driving inflammatory responses during disease development and progression.13-15 In 2020 alone, more than 3 million people in the US and European Union were diagnosed with IBD. Although the exact causes of IBD are unknown, pathobionts are believed to play a pivotal role in disease development.
THE ROLE OF IMMUNOGLOBULIN A
Immunoglobulin A (IgA) is the predominant antibody isotype secreted into the intestinal lumen, where it binds indigenous members of the microbiota and controls microbiota composition.16-19 IgA is also a critical mediator of intestinal immunity.20,21 Recognition of enteric pathogens by the intestinal immune system results in the production of high-affinity, T cell-dependent, pathogen-specific IgA, which is transcytosed into the intestinal lumen. In the lumen, these antibodies can bind and “coat” offending pathogens and provide protection against infection through neutralization and exclusion. Indigenous members of the intestinal microbiota also can stimulate IgA production and can become coated with IgA.22-24 While all intestinal bacteria can induce specific IgA responses, in principle, direct analyses of the proportion of intestinal bacteria that are coated with IgA demonstrated that only a fraction of all intestinal bacteria are measurably IgA coated.25-28 Because little is known about the specificity of the intestinal IgA response in the context of a complex microbiota, whether this fraction is composed of many species or a high percentage of a few species remains unclear. However, while several commensal bacteria have been shown to induce specific IgA responses, pathobionts and pathogens induce higher levels of IgA than “true” commensals.28 For example, SFB and Helicobacter species are potent inducers of IgA response in the intestine.29,30
The inflammasome is a critical component of the innate immune system that orchestrates the activation of Caspase-1 and release of the inflammatory cytokines IL-1β and IL-18 in response to infection or damage. Mice lacking components of the inflammasome, such as the signaling adaptor apoptosis-associated speck-like protein containing a CARD (ASC), harbor a dysbiotic microbiota that is colitogenic and can be transmitted to wild type mice through co-housing.31 In particular, acquisition of bacteria from the family Prevotellaceae has been implicated in colitis development in dysbiotic mice.32 Despite considerable effort, the identification of specific pathobionts responsible for driving the development of disease in humans has proven difficult due to the complexity and diversity of the microbiota, as well as the influence of host genetics and environment on disease susceptibility.
SUMMARY OF THE IGA-SEQ PATENTED INVENTION
IgA-SEQ capitalizes on the discovery that secretory antibodies can be used to detect and identify organisms present in the microbiota that influence susceptibility to or contribute to the development or progression of diseases or disorders, including chronic inflammatory diseases. However, compared to pathogen-induced IgA, commensal-induced IgA is generally believed to be of relatively low-affinity and specificity.33,34 Thus, relative levels of bacterial coating with IgA might be predicted to correlate with the magnitude of the inflammatory response triggered by a specific intestinal bacterial species. IgA-SEQ is a method for identifying the target organisms in the microbiota contributing to an inflammatory process and, by default, is on or near any mucosal surface of the patient including: the gastrointestinal, the respiratory, and genitourinary tracts. In addition to IBD, the role of pathobionts and chronic inflammation have been implicated in multiple conditions, including celiac disease, acute diverticulitis, intestinal hyperplasia, metabolic syndrome, obesity, rheumatoid arthritis, liver disease, hepatic steatosis, fatty liver disease, non-alcoholic fatty liver disease, and non-alcoholic steatohepatitis.
FLAGSHIP INFLAMMATORY BOWEL DISEASE PROGRAM UTILIZING IGA-SEQ™
Metagenomic studies comparing the microbiota of diseased and normal individuals have failed to identify disease-causing bacteria for a number of reasons, notwithstanding limited technical methodologies. There is a need in the field to not only identify bacteria in the microbiota that are implicated in the development or progression of certain diseases and disorders, but to determine which are activating the chronic inflammatory pathways and exactly how. When identifying promising microbiome therapeutic approaches for any disease or condition, it is critical to understand the mechanism of disease and disease pathways as well as the role of the microbiome. IgA−SEQ begins that discovery process whereby the human gut now becomes a crucial drug discovery tool. IgA−SEQ was developed at Yale, licensed by Artizan, and utilizes flow cytometry-based bacterial cell sorting and 16S sequencing to characterize taxa-specific coating of the intestinal microbiota with immunoglobulin A. It has been demonstrated that the high IgA−coating uniquely identifies colitogenic intestinal bacteria in a mouse model of microbiota-driven colitis. Proof of principle was shown by leveraging the IgA−SEQ methodology to identify culprit pathobionts and proceed with extensive anaerobic culturing of these organisms from patients with IBD to create personalized disease-associated gut microbiota culture collections with pre-defined levels of IgA coating. Using these collections, it was found that intestinal bacteria selected on the basis of high coating with IgA conferred significant susceptibility to colitis in germ-free mice. Thus, these studies suggested that IgA−coating identifies inflammatory commensals that preferentially drive intestinal disease.35 Inflammation within the intestine and distal organs can be mediated by innate and adaptive components of the immune response. IgA-SEQ is based on the rationale that certain intestinal bacteria that become coated with IgA are close to the host mucosal surfaces and are activating and promulgating a pro-inflammatory response. Thus, IgA-SEQ is a promising approach for identifying colitogenic bacteria in man. The influence of these gut microbiota/host immune interactions, however, is not limited to the gut. More recently, enrichment of IgA-coated microbiota has been demonstrated for Crohn’s disease associated spondylarthritis.36 Thus, IgA-SEQ might be applicable to the identification of microbial strains in the gut microbiota, which, through an inflammatory circuitry, are modulating inflammation and driving disease at distant sites.
The mammalian immune system has evolved in the presence of a complex community of indigenous microorganisms that constitutively colonize all barrier surfaces. This intimate relationship has resulted in the development of a vast array of reciprocal interactions between the microbiota and the host immune system, particularly in the intestine, where the density and diversity of indigenous microbes are greatest. The relationship between the immune system and the microbiota is central to the impact of the microbiota on disease, thus, immunologically important pathobionts are likely to have outsized effects on human disease. By utilizing the novel IgA-SEQ approach, Artizan’s team of researchers can take advantage of mucosal antibody responses to the microbiota to identify immunologically relevant microbes that may play causal roles in chronic disease.
PIONEERING TRANSFORMATIVE PRECISION THERAPEUTICS
Artizan utilizes IgA-SEQ and its world-class biobanking program with sophisticated bioinformatics system to enable its research team to examine microbial drivers in many different disease types. IgA-SEQ allows researchers to methodically investigate microbial communities down to individual bacterial strains from healthy and disease-affected samples from its exclusive biobanking program. Once a strain of interest is identified, Artizan applies its mechanistic research expertise to determine the root causes of pathology. Deciphering the precise pathogenic mechanisms allows the company to design transformative disease-modifying therapeutic agents, including novel small molecule and monoclonal antibody treatments. Artizan’s unique capabilities identify predictive biomarkers to target precision treatment to the right subset of patients.
To date, this process has allowed Artizan to develop transformative precision therapeutics that target and block the root causes of diverse, serious diseases triggered by intestinal inflammation including gut exclusive microbial metalloprotease inhibitors (GEMMI). These precision therapeutics include ARZC-001, a novel, oral, gut-restricted potent small molecule inhibitor for the treatment of IBD. The company anticipates ARZC-001 will enter the clinic by early 2023. Additionally, Artizan is advancing other candidates into the late preclinical stage for IBD, each distinctly different in chemical composition and the target that it is aiming to inhibit.
Beyond IBD, Artizan is also using IgA-SEQ to identify and characterize microbial drivers of disease within precise patient subsets in other areas, including certain cancers and gastrointestinal, metabolic, autoimmune, and neurodegenerative disease. The company has a Parkinson’s Disease therapeutic discovery and development program that is progressing. Artizan also continues to identify partners for therapeutic development in other disease states as well as concepts that improve the tolerability and viability of currently available treatments in areas including oncology.
REFERENCES
- Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011 Aug;23(4):473-80. doi: 10.1016/j.coi.2011.07.010. Epub 2011 Aug 17. PMID: 21856139; PMCID: PMC3426444.
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010 Jun 25;32(6):815-27. doi: 10.1016/j.immuni.2010.06.001. PMID: 20620945; PMCID: PMC2904693.
- Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012 Jun 6;4(137):137rv7. doi: 10.1126/scitranslmed.3004184. PMID: 22674557; PMCID: PMC5020897.
- Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011 Aug;23(4):473-80. doi: 10.1016/j.coi.2011.07.010. Epub 2011 Aug 17. PMID: 21856139; PMCID: PMC3426444.
- Hooper DU, Adair EC, Cardinale BJ, Byrnes JE, Hungate BA, Matulich KL, Gonzalez A, Duffy JE, Gamfeldt L, O’Connor MI. A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature. 2012 May 2;486(7401):105-8. doi: 10.1038/nature11118. PMID: 22678289.
- Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011 Oct 20;10(4):311-23. doi: 10.1016/j.chom.2011.10.004. PMID: 22018232; PMCID: PMC3202012.
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012 Sep 13;489(7415):220-30. doi: 10.1038/nature11550. PMID: 22972295; PMCID: PMC3577372.
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011 Jan 21;331(6015):337-41. doi: 10.1126/science.1198469. Epub 2010 Dec 23. PMID: 21205640; PMCID: PMC3969237.
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010 Jun 25;32(6):815-27. doi: 10.1016/j.immuni.2010.06.001. PMID: 20620945; PMCID: PMC2904693.
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009 Oct 30;139(3):485-98. doi: 10.1016/j.cell.2009.09.033. PMID: 19836068; PMCID: PMC2796826.
- Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D.. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci USA 108: 11548-11553.
- Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011 Oct 20;10(4):311-23. doi: 10.1016/j.chom.2011.10.004. PMID: 22018232; PMCID: PMC3202012.
- Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009 Nov 19;361(21):2066-78. doi: 10.1056/NEJMra0804647. PMID: 19923578; PMCID: PMC3491806.
- Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014 Mar 12;15(3):382-392. doi: 10.1016/j.chom.2014.02.005. PMID: 24629344; PMCID: PMC4059512.
- Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013 Oct;62(10):1505-10. doi: 10.1136/gutjnl-2012-303954. PMID: 24037875; PMCID: PMC3822528.
- Macpherson AJ, Geuking MB, Slack E, Hapfelmeier S, McCoy KD. The habitat, double life, citizenship, and forgetfulness of IgA. Immunol Rev. 2012 Jan;245(1):132-46. doi: 10.1111/j.1600-065X.2011.01072.x. PMID: 22168417.
- Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012 Dec;12(12):821-32. doi: 10.1038/nri3322. Epub 2012 Oct 29. PMID: 23103985.
- Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004 Feb 17;101(7):1981-6. doi: 10.1073/pnas.0307317101. Epub 2004 Feb 6. PMID: 14766966; PMCID: PMC357038.
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007 Nov 15;2(5):328-39. doi: 10.1016/j.chom.2007.09.013. PMID: 18005754.
- Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012 Dec;12(12):821-32. doi: 10.1038/nri3322. Epub 2012 Oct 29. PMID: 23103985.
- Slack E, Balmer ML, Fritz JH, Hapfelmeier S. Functional flexibility of intestinal IgA – broadening the fine line. Front Immunol. 2012 May 3;3:100. doi: 10.3389/fimmu.2012.00100. PMID: 22563329; PMCID: PMC3342566.
- Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012 Dec;12(12):821-32. doi: 10.1038/nri3322. Epub 2012 Oct 29. PMID: 23103985.
- Slack E, Balmer ML, Fritz JH, Hapfelmeier S. Functional flexibility of intestinal IgA – broadening the fine line. Front Immunol. 2012 May 3;3:100. doi: 10.3389/fimmu.2012.00100. PMID: 22563329; PMCID: PMC3342566.
- van der Waaij LA, Mesander G, Limburg PC, van der Waaij D. Direct flow cytometry of anaerobic bacteria in human feces. Cytometry. 1994 Jul 1;16(3):270-9. doi: 10.1002/cyto.990160312. PMID: 7924697.
- Tsuruta T, Inoue R, Nojima I, Tsukahara T, Hara H, Yajima T. The amount of secreted IgA may not determine the secretory IgA coating ratio of gastrointestinal bacteria. FEMS Immunol Med Microbiol. 2009 Jul;56(2):185-9. doi: 10.1111/j.1574-695X.2009.00568.x. Epub 2009 May 7. PMID: 19490128.
- van der Waaij LA, Limburg PC, Mesander G, van der Waaij D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut. 1996 Mar;38(3):348-54. doi: 10.1136/gut.38.3.348. PMID: 8675085; PMCID: PMC1383061.
- van der Waaij LA, Mesander G, Limburg PC, van der Waaij D. Direct flow cytometry of anaerobic bacteria in human feces. Cytometry. 1994 Jul 1;16(3):270-9. doi: 10.1002/cyto.990160312. PMID: 7924697.
- Slack E, Balmer ML, Fritz JH, Hapfelmeier S. Functional flexibility of intestinal IgA – broadening the fine line. Front Immunol. 2012 May 3;3:100. doi: 10.3389/fimmu.2012.00100. PMID: 22563329; PMCID: PMC3342566.
- Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun. 1999 Jul;67(7):3504-11. doi: 10.1128/IAI.67.7.3504-3511.1999. PMID: 10377132; PMCID: PMC116537.
- Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999 Apr;67(4):1992-2000. doi: 10.1128/IAI.67.4.1992-2000.1999. PMID: 10085047; PMCID: PMC96557.
- Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012 Jan 18;481(7381):278-86. doi: 10.1038/nature10759. PMID: 22258606.
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011 May 27;145(5):745-57. doi: 10.1016/j.cell.2011.04.022. Epub 2011 May 12. PMID: 21565393; PMCID: PMC3140910.
- Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. 2012 Dec;12(12):821-32. doi: 10.1038/nri3322. Epub 2012 Oct 29. PMID: 23103985.
- Slack E, Balmer ML, Fritz JH, Hapfelmeier S. Functional flexibility of intestinal IgA – broadening the fine line. Front Immunol. 2012 May 3;3:100. doi: 10.3389/fimmu.2012.00100. PMID: 22563329; PMCID: PMC3342566.
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, Ruggiero E, Cho JH, Goodman AL, Flavell RA. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014 Aug 28;158(5):1000-1010. doi: 10.1016/j.cell.2014.08.006. PMID: 25171403; PMCID: PMC4174347.
- Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG, Woo V, Teng F, Tran NL, Sczesnak A, Chai C, Kim M, Diehl GE, Ajami NJ, Petrosino JF, Zhou XK, Schwartzman S, Mandl LA, Abramowitz M, Jacob V, Bosworth B, Steinlauf A, Scherl EJ, Wu HJ, Simpson KW, Longman RS. IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med. 2017 Feb 8;9(376):eaaf9655. doi: 10.1126/scitranslmed.aaf9655. PMID: 28179509; PMCID: PMC6159892.

Dr. Bridget Ann Martell is President and Chief Executive Officer of Artizan Biosciences. She is a seasoned biopharmaceutical executive with high-impact, results-oriented breadth and depth in clinical development, medical affairs, and business development across a range of therapeutic areas from first-in-human through approval and commercialization. She has held leadership and C-suite executive roles at companies, including Kura Oncology, where she led the Menin Inhibitor Program, and at Juniper Pharmaceuticals, where she led the development program for a Bob Langer-created drug/device technology that was out-licensed. She started her pharmaceutical career at Pfizer, where she had roles of increasing responsibility that included Clinical Team Leader for Sutent® GU solid tumors in the Oncology Business Unit and, ultimately, Biosimilars Medical Head. She began her clinical investigation career at Yale with a Career Development Award, where she led a therapeutic cocaine vaccine trial and was a Robert Wood Johnson Clinical Scholar. She was a practicing physician at Yale for 20 years and remains active as an Entrepreneur in Residence at the Yale Innovations Office. She earned her BSc in Microbiology from Cornell University, her MA in Molecular Immunology from Boston University, and her MD from The Chicago Medical School. Dr. Martell is board certified in Internal and Addiction Medicine.
Total Page Views: 2835