Issue:May 2020
DRUG DISCOVERY - Disrupting Drug Discovery From Assay Development to Lead Compound
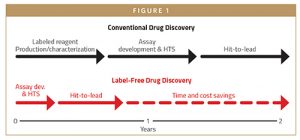
A PATH FOR DRUG DISCOVERY
High-throughput screening remains one of the most powerful, unbiased approaches for small molecule drug discovery. Compound libraries, many featuring millions of diverse molecules, are rapidly tested against a therapeutic target to identify the most promising leads for preclinical and clinical development.
Today, the high-throughput screening toolbox features traditional label-dependent approaches and novel label-free technologies. Researchers select the most promising methodology based on assay sensitivity, data quality, costs, and speed. Moreover, many companies outsource drug discovery to contract research organizations (CROs) with expertise and an established infrastructure to manage hit identification and hit-to-lead programs.
IDENTIFYING THE RIGHT ASSAY FOR YOUR TARGET
When a target has been validated for drug discovery, the first step is to develop a robust assay suitable for measuring its activity and revealing small molecule modulators. Assay development aims to address:
-How sensitive and robust is the assay?
-What is the linear range of the enzyme activity?
-What are the kinetic parameters to achieve balanced conditions?
Every high-throughput assay methodology claims to provide answers and a sensitive and reproducible assay suitable for hit identification. However, before selection, two additional questions apply:
-What limitations and/or restrictions could impact data quality?
-Will extensive false-positive and/or false-negative results delay progress and increase costs?
Answering these questions is paramount to foresee potential obstacles and to determine measures of success for a drug discovery program.
Let’s explore these questions in the context of different approaches: label-dependent, emerging label-free, and self-assembled monolayer desorption ionization label-free formats.
DON’T BELIEVE IN LABELS, BELIEVE IN RESULTS
Label-dependent assays are prevalent in drug discovery. They measure a change in radioactivity or optical readouts, such as fluorescence or luminescence. Popular optical-based assays take advantage of energy transfer between a donor and acceptor reagent, such as a bead or fluorescent probe.
These formats are generally simple and inexpensive to run. However, possible limitations and significant opportunities for false-positive data make them potentially problematic.
LABEL-FREE ASSAYS: SEEING IS BELIEVING, OR IS IT?
Label-free assays eliminate the false-positive and false-negative results that derive from optical interference of library compounds. This advantage alone can expedite drug discovery by avoiding the need to rule out false positives through orthogonal assays. Mass spectrometry (MS) has emerged as a powerful, label-free readout to analyze biochemical reactions. MS reveals the precise mass of substrates and products, providing an information-rich, quantitative readout that eliminates questions surrounding the data. This also opens avenues to identify unanticipated reactions.
Electrospray Ionization Mass Spectrometry
Conventional MS instruments, such as electrospray ionization, offer high sensitivity well suited for analyzing biochemical reactions. However, reactions require processing often by solid-phase extraction prior to analysis. This time-consuming step raises significant limitations for screening applications:
-Costly consumables are needed.
-Required reaction volumes may be 10+ times higher than other formats, driving up reagent consumption and costs.
-Not amenable to 1536 geometries.
-Low throughput: a 384-well plate takes over an hour to read.
Acoustic Dispense With Mass Spectrometry
An emerging strategy integrates acoustic transfer with MS.2 Directly injecting biochemical reactions into the MS instrument eliminates the rate-limiting sample preparation step. Prototype instruments achieve a throughput of 100,000 samples per day, useful to researchers screening millions of compounds.2 Interestingly, what enables the rapidity introduces potential limitations:
-Critical reaction components — salts, detergents, and carrier proteins — lead to ion suppression.
-Ion suppression introduces noise, and data quality suffers, with Z’-factors falling below 0.5.2
-Certain reaction components may impact MS hardware, necessitating frequent maintenance.
-Low-sensitivity challenges detection of nanomolar analytes.
As a solution, researchers may exclude the salts, detergents, and carrier proteins. However, this can impact enzyme activity and aggregation of proteins or screening compounds. At low or zero salt concentrations, can activity be measured? Is the assay sensitive enough to work under balanced conditions (at substrate Km)? Commercial instruments integrating acoustic dispense with an open port interface along with MS are expected in 2020, and while preliminary data is encouraging and eliminates the tedious sample preparation steps, it remains to be understood whether inherent restrictions with ESI MS limit wide scale drug discovery applications.
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry
An alternative to electrospray ionization, MALDI-TOF MS generates a rapid readout useful for high-throughput applications. It measures diverse analytes, including small molecules, peptides, oligonucleotides, and proteins (including antibodies). MALDI samples are prepared by introducing a matrix molecule to the analyte to assist with ionization to enable detection. As with acoustic MS, ion suppression due to salts, detergents, and carrier proteins is a significant limitation. These components must be removed or excluded prior to analysis, resulting in the same issues as for acoustic MS. Finding a buffer system that both the enzyme and instrumentation can tolerate is time-consuming: Some researchers report yearlong assay development programs.
Considering the aforementioned options for drug discovery, researchers must still decide which matters more: biochemical relevance or speed and take time to validate and further test their results through orthogonal approaches.
A DISRUPTIVE LABEL-FREE SOLUTION FOR SUCCESSFUL DRUG DISCOVERY
Self-assembled monolayers for matrix-assisted laser desorption/ionization — a technique termed SAMDI — was first reported in 2002.3 In 2011, this innovative assay methodology had already achieved a throughput capable of screening 100,000 compounds in 1 day and it continues to evolve with proven assays for high-throughput screening of diverse targets and biochemical activities, affinity selection assays, and cell-based assays.4-7
A significant advantage of this unique surface chemistry is that drug discovery researchers are free to optimize assay conditions according to their targets, rather than having the analytical method dictate how to run their reactions. The result is more biochemically relevant assay conditions, more relevant data, and higher confidence
in results. These key innovations accelerate assay development, hit identification with high-throughput screening, and hit-to-lead optimization.
SURFACE CHEMISTRY: LAYING A FOUNDATION FOR SUCCESS
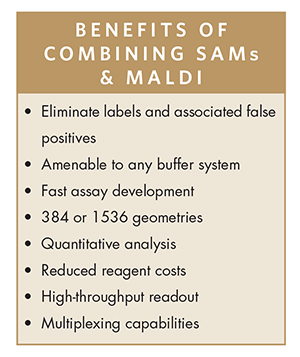
Self-assembled monolayers (SAMs) of alkanethiolates on gold offer a structurally well-defined and synthetically flexible surface that enables key advantages for biochemical reactions:
-SAMs presenting short oligomers of ethylene glycol are inert to non-specific protein adsorption.8
-SAMs can be tailored with functional groups to specifically immobilize distinct ligands, including small molecules, peptides, oligonucleotides, proteins, and antibodies. Other reaction components are washed away, purifying up to 1,536 samples in seconds and eliminating ion suppression.
-SAMs are efficient substrates for MALDITOF MS, the basis for the ultra-high-throughput and label-free readout.
The assay platform is simple to utilize: The assays start with a homogenous biochemical reaction in a 384- or 1535-well plate. When the reactions are quenched, complex reaction mixtures are transferred to high-density 384 or 1536 biochip arrays. There, substrates and products are specifically captured to the SAM, while other reaction components are washed away. This innovation enables researchers to include any salt, buffer, detergent, carrier protein, or other component in the biochemical reaction.
Following immobilization of the specific analytes, matrix is applied to the biochip array and analyzed in a MALDI instrument. The resulting spectra are clean, with peaks corresponding primarily to singly charged species with virtually no fragmentation for simple data interpretation and minimal background.
ASSAY DEVELOPMENT: SMARTER QUESTIONS, FASTER ANSWERS
Assay development with self-assembled monolayer desorption/ionization starts ahead of other assay formats: There is no need to synthesize fluorescent reporters, raise antibodies, or optimize a compatible buffer. Regardless of the analyte, assays that reveal balanced conditions suitable for hit identification and characterization can be developed in days.
One aspect to consider is that the analyte of interest must be amenable to immobilization on the SAM array. For this, several immobilization chemistries have been established, with new, traceless innovations opening avenues to immobilize challenging molecules.9
DISRUPTING THE HIGHTHROUGHPUT SCREENING MARKET
Researchers can now be assured that speed AND biological relevance are not compromised when running biochemical assays. The use of MALDI TOF MS provides an ultra-high-throughput readout, with the newest instruments reading a 1536-spot plate in less than 10 minutes. Meanwhile, the self-assembled monolayers provide a flexible platform for virtually any buffer, any analyte, and any target, making this combination the gold standard for drug discovery.
Another innovation is the ability to multiplex assays. Not only can this platform distinguish multiple products from a single enzyme, but it can also be used to measure multiple activities from distinct enzymes. Provided the substrates and products are mass resolvable, the reactions can be run with two or more enzymes in a single well, or run in separate plates and multiplexed prior to immobilization and analysis. In this manner, twice the data is generated in half the time. The method can also shed light on compound potency and selectivity simultaneously by screening compounds against different targets.
HIT-TO-LEAD: AN OPTIMIZED ASSAY GENERATES OPTIMIZED RESULTS
When initial hits have been identified, revealing structure-activity relationships (SAR) and optimizing for potency and selectivity are simple and streamlined. The clean dose-response data resulting from the self-assembled monolayers and MALDI MS drives medicinal chemistry programs. The platform’s flexibility enables experiments that elucidate mechanism of action, defining competitive, uncompetitive, noncompetitive, or mixed inhibition. This innovation allows total characterization of lead compounds from hit identification to inhibition mechanism and generates data to drive decisions for further development.
BINDING ASSAYS: RAPID, LABEL-FREE SOLUTIONS FOR CHALLENGING PROBLEMS
Many drug discovery programs seek small molecules that bind a specific target. When binding does not change functional activity or the target itself does not possess enzymatic activity, binding assays must be developed to reveal small molecules that engage a therapeutic target of interest. Long-established binding assay types include low-throughput biophysical approaches such as surface plasmon resonance and isothermal calorimetry.
Affinity selection mass spectrometry has emerged as a powerful tool for revealing non-covalent small molecule binders. It informs on the compound mass rather than on a signal change, where metabolites or impurities may generate false-positive results. Conventional MS binding assays utilize chromatography to isolate a target and bound small molecule from non-binders. A second column dissociates the small molecule from the target. Finally, the small molecule is analyzed by MS. This approach is suited for measuring binding events, provided that the target is stable throughout the protocol.
In addition, requisite high target concentrations (often > 1 μM), increase costs — a challenge for difficult to express/synthesize targets. Lastly, to increase throughput, hundreds of compounds are often pooled in a single well. Mixing so many compounds in a single reaction may impact target and compound behavior. Alternatively, innovation with self-assembled monolayers has opened avenues to ultra-high-throughput, label-free assays that reveal non-covalent small molecule binders for diverse analytes. After the target and one or a pool of small molecules incubate, target-compound complexes are rapidly and selectively immobilized on the SAM-presenting biochip arrays and then directly analyzed to identify the binding molecules. Importantly, the monolayer molecule itself, at a constant density across the arrays, provides an internal standard to compare mass accuracy and binding across large-scale screens. Data analysis includes comparing the area under the curve (AUC) of any binding compound to the AUC of the monolayer molecule. This rapid analysis identifies potential binders. Dose response experiments rank-order the binding molecules to guide development efforts.6
SUMMARY
For many years, drug discovery researchers have been forced to compromise certain scientific assay goals — sacrificing data quality and costs to achieve high-throughput-labeled assays, sacrificing speed to use biochemically relevant reaction conditions with slow label-free readouts, or sacrificing optimal assay conditions to use rapid label-free readouts. With new innovations in assay methodologies, scientists no longer have to make sacrifices to do quality drug discovery research. The combination of high-density self-assembled monolayer biochip arrays and mass spectrometry disrupts the market by allowing researchers to use optimal reaction conditions with a label-free and ultra-high-throughput readout. This solution accelerates assay development, hit identification, and hit-to-lead characterization, taking discovery research one step closer to the clinic.
REFERENCES
- Kaeberlein M, McDonagh T, Heltweg B, et al. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 2005;280:17038-45.
- Sinclair I, Bachman M, Addison D, et al. Acoustic Mist Ionization Platform for Direct and Contactless Ultrahigh-Throughput Mass Spectrometry Analysis of Liquid Samples. Anal Chem. 2019;91:3790-4. DeLand P. Accelerating Assay Development and Prosecution, 2018 Technote.
- Su J & Mrksich M. Using mass spectrometry to characterize self-assembled monolayers presenting peptides, proteins, and carbohydrates. Angew Chem Int Ed. 2002;41:4715-8.
- Gurard-Levin ZA, Scholle MD, Eisenberg AH, et al. High-throughput screening of small molecule libraries using SAMDI mass spectrometry. ACS Comb Sci. 2011;13:347-50.
- Gurard-Levin ZA & Mrksich, M. Combining self-assembled monolayers and mass spectrometry for applications in biochips. Annu Rev Anal Chem. 2008;1:767-800.
- VanderPorten EC, Scholle MD, Sherrill J, et al. Identification of small molecule non-covalent binders utilizing SAMDI technology. SLAS Discov. 2017;22:1211-7.
- Berns EJ, Cabezas MD, Mrksich M. Cellular assays with a molecular endpoint measured by SAMDI mass spectrometry. Small. 2016;12:3811-8.
- Mrksich M & Whitesides GM. Using self-assembled monolayers that present oligo (ethylene glycol) groups to control the interactions of proteins with surfaces. In Poly(ethylene glycol) Chemistry and Biological Applications, 1997 ACS Symposium Series. 680:361-73.
- Helal KY, Alamgir A, Berns EJ, et al. Traceless immobilization of analytes for high-throughput experiments with SAMDI mass spectrometry. JACS. 2018;140:8060.
To view this issue and all back issues online, please visit www.drug-dev.com.

Dr. Zack Gurard-Levin has served as Chief Scientific Officer at SAMDI Tech, Inc. since June 2016. He brings more than 10 years of multidisciplinary research experience with expertise in chemistry, biochemistry, cellular biology, and translational research. Dr. Gurard-Levin was a pioneer user of SAMDI technology and co-developed SAMDI as a high-throughput, label-free solution for drug discovery research. Prior to SAMDI Tech, he was a research scientist at the Institut Curie in Paris, France, leading epigenetics drug discovery and diagnostics projects in oncology. He has authored numerous peer-reviewed articles and has been awarded multiple research grants. He earned his PhD in Chemistry from the University of Chicago and completed a post-doctoral fellowship at Institut Curie.
Total Page Views: 4595