Issue:January/February 2023
DISCOVERY PLATFORM - Reverse-Engineering Targeted Immunotherapy
THE MODERN ERA OF CANCER IMMUNOTHERAPY
Cancer therapy has undergone a profound change over the recent past with the introduction of successful therapies that specifically target the human immune system. Cancer immunotherapy originally dates to 1891 with the observation that admixtures of two bacteria could cause regression in patients with sarcoma.1 But it took about 100 years for the FDA to approve the first drugs specifically targeting the human immune system for the treatment of cancer with the approvals of interleukin-2 and interferon-alpha in the 1990s. These two cytokine products activate lymphoid and other cells in a fairly non-specific manner, and the high doses administered often lead to severe complications and require administration in specialized settings. The modern era of targeted immunotherapy was ushered in with the approval of the first T-cell checkpoint inhibitor (CPI), the anti-CTLA-4 agent ipilimumab, in 2011 for the treatment of metastatic melanoma. This was quickly followed by approvals for antagonists of PD-1 and PD-L1, the two key molecules important for regulation of T cells within the tumor. With these approvals, clinicians now had agents that targeted specific components of the immune system that could induce activated T cells to attack cancers.
The incorporation of CPIs into standard therapy has significantly changed the cancer treatment landscape. Used alone, the best observed response rates (that is reduction in tumor volumes) hover around 35%, and while many patients are living longer, most are not cured of their disease. Much has been learned about the intrinsic determinants of response or resistance, centered on the understanding that tumors that have an already active immune tumor microenvironment (TME), with active infiltration of T cells (so-called “hot” tumors) are most likely to respond, whereas those that lack such infiltration (so-called “cold” tumors) are less so.2 Central to the immunosuppression within the TME are tumor-associated macrophages (TAMs), which produce cytokines and other factors that foster an immunosuppressive environment and prevent the full activation of T cells following CPI therapy. OncoResponse set out to use our proprietary technology to identify targets on TAMs that could be manipulated to reverse this immunosuppression and promote the immune system’s ability to efficiently kill tumor cells and enable clinical responses.
FOCUSING ON “ELITE RESPONDERS” TO CANCER IMMUNOTHERAPY
Our hypothesis was that patients who have exquisite responses to CPI therapy — we call these patients Elite Responders — have autoantibodies that help activate other components of the immune system and potentiate their response to CPIs. It is well known that cancer patients have a reduced self-tolerance and can produce autoantibodies to self-antigens. We thought to use this disruption of normal immune tolerance to identify antibodies made to normal proteins that might have a regulatory or immunostimulatory effect. In particular, we focused on TAMs, those key regulators of immunosuppression in the TME, looking for autoantibodies made by Elite Responders that could help reverse the immunosuppressive activities of these cells.
Macrophages in the TME exist in two broad categories: immunoinhibitory (called M2 macrophages) macrophages and immunostimulatory (or M1) macrophages. The majority of TAMs are M2 macrophages, which leads to immunosuppression in the TME. Using samples from Elite Responders, we looked for antibodies that bound to M2 macrophages, but did not bind M1. The ultimate goal was to identify an antibody that had a regulatory effect on TAMs that could reprogram them to be more M1-like.
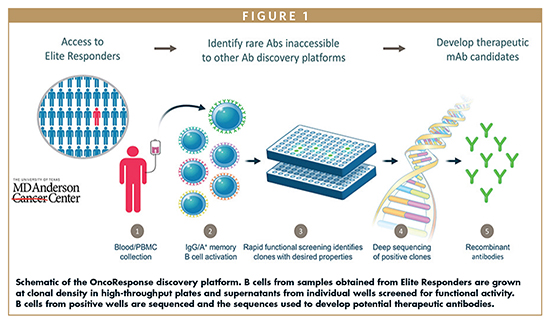
One of the most difficult things to do in oncology drug development is obtain samples from patients with a specific treatment history outside of a sponsored clinical trial. When OncoResponse formulated the strategy to look for rare regulatory antibodies using our platform, it quickly became clear we would need a reliable source of high-quality samples, in particular serum and whole blood, to screen for potential hits. Our best chance of finding interesting autoantibodies would be to use samples from patients who had had exquisite responses to CPI therapy. So, we sought samples from patients who had partial or complete tumor reduction that lasted a minimum of 3 months. The key breakthrough came when we partnered with MD Anderson Cancer Center to identify such patients and rapidly obtain the necessary samples. We were able to negotiate a collaborative agreement that gave us access to numerous patient samples with minimal regulatory hurdles. Through this collaborative framework, we were able to screen sufficient samples for our lead generation.
A NEW APPROACH TO DISCOVER CANCER THERAPEUTICS
Human memory B cells are specialized lymphocytes that produce antibodies. Each B cell produces a monoclonal antibody to a specific antigen. Identifying a B cell that produces a particular antibody allows identification of the sequence of that antibody and subsequent production in a laboratory or commercial setting. OncoResponse’s proprietary technology allows us to culture human B cells at clonal density, allowing identification of antibodies that are produced by a particular memory B cell. We can use the supernatants (ie, the monoclonal antibodies) from these cultures to screen against a protein or cell type of interest, looking for those B cells that produce antibodies that might modulate the response to cancer immunotherapies.
We began by screening serum from Elite Responders looking for antibodies that bound to M2 TAMs. B-cell cloning and production is resource intensive and first screening serum from Elite Responders against the selected target allowed us to narrow down the number of patients from whose blood we needed to grow B cells. After identification of patient serum samples that were positive hits, that is, contained pools of antibodies that bound to M2 macrophages, we went back to whole blood samples from these patients and cultured B cells for subsequent screening in the same assay. B cells were grown at clonal density, and supernatants from these cultures were then used for screens for activity. We confirmed binding to M2 macrophages and lack of binding to M1 macrophages, and also looked for functional activity. One of the key advantages of the OncoResponse platform is the ability to identify functionally active antibodies early in the screen. Using the appropriate functional screen, in this case the ability to repolarize M2 macrophages to a more M1-like phenotype, we were able to identify antibodies with appropriate functional characteristics before we even knew the target of the antibody. In this way, we could reverse engineer an antibody that we know already has potential functional characteristics derived from a patient who experienced an exquisite response. Presumably, such antibodies were either present before treatment or were newly generated following the initiation of therapy. While we first targeted M2 macrophages in this screening campaign, we can use the same technology to look for targets on other cell types that may regulate responses to therapy.
Following identification of the appropriate B cell clone, deep sequencing can identify the sequence of the target antibody. In the case of multiple clones in one well, that is that more than one memory B cell was present in the original well, further experimental validation is required to identify the proper clone. To do this, we deep sequence the immunoglobulin heavy and light chain sequences and engineer the possible combinations. We can then test these in our functional assays to identify the correct combination that mimics the effects we saw in the original assays. After the right sequence is identified, cloning into a consensus vector for production and further characterization allows for rapid characterization.
A salient feature of this process is that functional antibodies, which might not be identified through conventional antibody development methods, are rapidly chosen. A hypothetical example of the advantage of this system may be seen if looking for an antibody that acts as an agonist to a receptor. In our functional screening technology, only antibodies that are activating for the receptor would be identified. On the other hand, conventional phage display or murine technologies will produce antibodies against any antigenic epitope, which must then be screened. If the activated conformation of the receptor is not easily replicated in the screening system, it may be impossible to identify an activating antibody. Similarly, if the epitope to be targeted is poorly immunogenic, or masked, or requires a conformational change, it may not be identified through conventional techniques. Because we screen for functionally active antibodies, we can identify and clone antibodies to unique epitopes that are immediately relevant and not easily amenable to targeting by other methods.
TAKING THE RESPONSE BACK TO PATIENTS
OR2805 was cloned from our original screen of Elite Responders looking for antibodies that targeted M2 macrophages. Biochemical and biophysical experiments identified the target of OR2805 as CD163.3 At first blush, CD163 is not an obvious target. CD163 is a well-known macrophage scavenger receptor that has as its normal role to bind the hemoglobin-haptoglobin complex and clear this from circulation. CD163 is known to be upregulated on M2 macrophages and is considered a marker for such macrophages in the TME. OR2805 was found to bind to CD163 simultaneously in two separate domains of the molecule and likely induces a conformational change in the molecule leading to downstream effects that fundamentally reorient the functional status of the macrophage. This surprising observation suggests that the mechanism of action is not simply blockade of normal ligand binding, but a new and unique function of CD163 that could only have been identified by a functional screen such as the one we performed.
Extensive characterization of the interaction of OR2805 with CD163 demonstrates that it can potently reverse the phenotype of M2 macrophages. We have demonstrated that incubating M2 macrophages with OR2805 causes them to decrease expression of cell surface markers for M2 macrophages and increase those for M1 macrophages. The most important data, however, are the effects that OR2805 has on T-cell activation by macrophages. When T cells and M1 macrophages are co-cultured and the T cells stimulated by activation of the CD3 receptor, markers of activation such as the cytokines IL-2 and interferon gamma are produced and the T cells proliferate. In the presence of M2 macrophages, this activation and proliferation are prevented. When M2 macrophages are treated with OR2805, however, the phenotype is reversed, and robust activation and proliferation are observed. Finally, we have tested OR2805 in a mouse models of lung cancer. In these models, we used pembrolizumab (which is marketed for treatment of certain cancers as Keytruda®) as the gold standard for comparison. OR2805 demonstrated anti-tumor activity in these models that was equal or superior to that observed with pembrolizumab. Taken together, these data formed the basis of our IND and entry into the clinic.
OR2805 is now in Phase 1 clinical testing at multiple sites in the US. This represents the full circle of discovery and development, from the patient to the lab and back into clinical testing. The ability to identify functionally important antibodies produced by cancer patients responding to CPI therapy is a novel and powerful tool for the development of new therapeutics. While OR2805 is the first such therapy to enter clinical testing, no doubt other antibodies that can potentiate the response of standard-of-care therapies or induce responses on their own await discovery. Although macrophages play an important role in immunosuppression in the TME, other cell types contribute, and novel targets and mechanisms of action also await discovery. By applying our patient-first technology and looking for clues in the immune system, we can harness a powerful tool — the human body — to identify and produce new molecules to make medicines for cancer patients.
REFERENCES
- The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications Zhang, Yuanyuan; Zhang, Zemin Cellular & Molecular Immunology 17:8, 807-821 DOI:10.1038/s41423-020-0488-6.
- Cancer immunotherapy using checkpoint blockade Ribas, Antoni; Wolchok, Jedd D., Science, 359: 6382, 1350-1355. DOI: 10.1126/science.aar4060.
- Targeting of CD163+ Macrophages in Inflammatory and Malignant Diseases. Skytthe, Maria K; Graversen, Jonas Heilskov; Moestrup, Søren K, International Journal of Molecular Sciences, 21:15 DOI:10.3390/ijms21155497.

Dr. Bob Lechleider joined OncoResponse from Seattle Genetics, where he was Senior Vice President of Clinical Development and was responsible for directing development of the early and late-stage portfolios, including the successful initial licensing of Padcev for the treatment of previously treated metastatic urothelial cancer. Prior to his work at Seattle Genetics, he held key roles with increasing responsibility at several biopharmaceutical companies, including MacroGenics, Human Genome Sciences, and MedImmune/Astra Zeneca. Before joining the biotechnology industry, he served as Assistant Professor of Pharmacology at the Uniformed Services University of the Health Sciences, and Associate Professor of Cell Biology at Georgetown University Medical School. He earned his AB cum laude from Princeton University and his MD from the University of Illinois College of Medicine at Chicago. He received clinical training in internal medicine at Beth Israel-Deaconess Medical Center in Boston and in medical oncology at the National Cancer Institute in Bethesda. He was also a Howard Hughes Medical Institute Scholar and a Damon Runyon-Walter Winchell postdoctoral fellow.
Total Page Views: 2268