Issue:April 2024
CANCER BIOMARKERS - Co-Analysis of CTCs & ctDNA: Gaining Multi-Dimensional Insights Into Cancer Heterogeneity
INTRODUCTION
Despite the progress made in cancer diagnosis and treatment, metastasis remains the predominant cause of cancer-related fatalities worldwide. A significant constraint to enhancing patient outcomes lies in the limitations of current clinical diagnostic tools, which struggle to predict the progression of metastasis and detect minimal residual disease, ie, a small number of cancer cells that are often undetectable by conventional methods post-therapy. Standard-of-care (SOC) methods, which include tissue biopsies, imaging technologies, and tumor markers, often fall short in assessing comprehensively all aspects of a patient’s cancer.1 This limitation arises from diagnostic thresholds that are not robust, sampling biases associated with temporal and spatial tumor heterogeneity, and constrained access to metastatic lesions.
A promising solution emerges in the form of liquid biopsies, offering a real-time and minimally invasive avenue for early detection, prognosis, solid tumor status monitoring, and predicting responses to cancer therapy.2 The concept of liquid biopsy based on circulating tumor cell (CTC) detection was proposed in 2011, followed by the inclusion of circulating tumor DNA (ctDNA) utilization for obtaining tumor biological information within the scope of liquid biopsy.3,4
With significant translational potential, numerous clinical trials are currently leveraging CTCs and ctDNA as indicators of tumor burden and progression, each reflecting distinct biological aspects of the disease.2 Each component presents unique advantages, with both CTCs and ctDNA providing complementary information about tumor dynamics. ctDNA, small cell-free DNA fragments (~160bp) present in liquid biopsy samples, has garnered more attention from researchers due to its ease of isolation from blood and the presence of cancer biomarkers-of-interest that have the potential to impact therapy decisions.5 In contrast, CTCs, though typically sparce in blood samples (as few as 1 cell/mL of blood), offer a more comprehensive source of genomic, gene expression, and epigenetic gene regulation information from living intact cells. CTC analysis can provide insights into various aspects of cancer biology and other characteristics associated with metastatic behavior, including endothelial-to-mesenchymal transition (EMT) changes responsible for CTC loss of adhesion from the primary tumor and entry into the circulatory system to eventually form secondary tumors in distant organs.6
Co-analysis of CTC abundance and phenotypic changes together with ctDNA concentration could allow for real-time monitoring of disease progression and reoccurrence, while genetic and epigenetic changes in CTCs and ctDNA mutations over time could provide valuable insights into the effectiveness of therapeutic interventions, as the presence of different somatic mutations may indicate cancer susceptibility or resistance to certain treatments (Figure 1).7 Such proposed dynamic monitoring could guide adjustments to treatment strategies, ensuring a more personalized approach to cancer treatment.
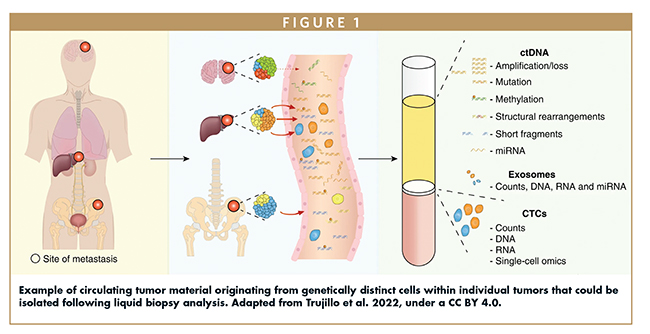
UNDERSTANDING THE LINK BETWEEN TUMOR HETEROGENEITY & TREATMENT EFFICACY
Cancer therapies depend, to a large extent, upon our ability to capture cancer tumor heterogeneity, ie, differences in their DNA and gene expression, which allow us to predict the tumor’s susceptibility or resistance to specific drug or hormone therapies. Monitoring phenotypic, genetic, and epigenetic CTC changes, and ctDNA mutations over time, could provide valuable insights into the effectiveness of therapeutic interventions. This dynamic monitoring could prove useful to guide adjustments to treatment strategies, ensuring more precise and targeted interventions.
CTCs, as a diverse cell population with distinct molecular phenotypes from primary tumors, represent an accessible and up-to-date cancer sample for real-time monitoring and detection of diagnostic biomarkers and therapeutic targets. In addition to their association with metastasis, CTCs can possess DNA mutations and unique surface markers not found in the primary tumor that can reduce the efficacy of radiation and targeted therapies tailored to the primary tumor. For example, HER2 expression on CTCs captured from breast cancer (BC) patients is often associated with unfavorable clinical outcomes and poor response to anti-HER2 therapy. This expression may represent a change in biomarker phenotype from the primary tumor from which the CTC was derived (ie, HER2 negative primary tumor).6
The impact of ctDNA analysis on clinical outcomes is evident in certain cancer types, such as non-small cell lung cancer (NSCLC) and BC.6 For example, higher baseline EGFR mutation levels derived from the analysis of ctDNA from NSCLC patients has been associated with increased tumor growth rates.8,9 In general, an increase in ctDNA levels can serve as an indicator of tumor burden, though the short half-life of ctDNA in circulation prevents the establishment of a specific cut-off concentration value for diagnosing cancer.9 Simultaneously, surpassing a defined threshold of CTC numbers becomes a clear indicator of the patient’s disease status, which may offer valuable insights into prognosis and survival.
An integrated approach, involving measurement of ctDNA and CTC levels combined with mutation profiling, could contribute to a more comprehensive understanding of tumor heterogeneity, facilitating more informed and personalized cancer treatment decisions.10
ADVANCES IN THE DETECTION OF CTCS & CTDNA
Integration of CTCs into clinical practice as a liquid biopsy analyte will require unbiased, efficient, rapid, and cost-effective capture technologies capable of isolating consistently a sufficient quantity of these rare cells. These capture technologies must also integrate seamlessly with advanced sequencing or quantitative tools and functional assays to produce data essential for accurate therapeutic decision-making. However, the complexity lies in the rarity and phenotypic diversity of CTCs, posing significant challenges for cell enumeration and characterization. As such, efficient CTC enrichment is crucial for reproducible downstream analysis and applications.
In recent years, technological advances have paved the way for the development of highly efficient and automatable techniques used to capture and enrich rare CTCs from liquid biopsy samples. Commercial instruments have emerged as a result of these advances, broadly utilizing antigen-dependent and antigen-independent approaches. Typically, antibody-based CTC capture processes involve selective CTC capture utilizing biomarkers that are highly expressed on CTCs and minimally expressed on other circulating cells, such as the Epithelial Cell Adhesion (EpCAM) biomarker. Often, this approach is combined with the negative selection of normal blood leukocytes cells using CD45-based methods to enhance assay specificity.6 However, CTCs transitioning to the mesenchymal state may exhibit low expression of these markers, potentially resulting in them being undetected. Assays capable of detecting multiple biomarkers indicative of cancer progression and metastasis are essential. As such, single-cell multi-omics analysis, coupled with long-term follow-up data, can unveil biomarkers associated with disease progression.11
On the other hand, antigen-agnostic detection technologies exploit physical properties such as size, charge, density, or elasticity for the enrichment of CTCs, overcoming challenges associated with biomarker expression. Various methods, including filter-based devices, density gradient centrifugation, capture surfaces, and microfluidic systems employ physical characteristics for CTC detection.1 Some instruments employ size-based selection of CTCs, which capture cells that are in the CTC size range of 8-30 microns and eliminates most white and red blood cells which are less than 8 microns in size. The size-based capture approach holds a notable advantage in not relying on biomarker expression, that is, even CTCs with varying or alternative biomarker expression profiles can be sorted and collected by size. Some innovative instruments offer a dual functionality of CTC capture and immunostain-based enumeration within a single workflow, all conducted on a microfluidic slide.12 Microfluidic platforms can also integrate biomarker-based positive and negative selection (eg, EpCAM, EGFR, HER2, and CD45), imaging, and micromanipulation to isolate pure subsets of CTCs. Advances in capture technologies are enabling detailed molecular and functional analyses of CTCs, at both bulk and single-cell levels, moving beyond simple enumeration. Combined with drug phenotyping, single-cell analysis of individual CTCs and CTC clusters can be utilized for the identification of potential therapeutic targets.1 While CTC capture and downstream analysis may show clinical relevance, many techniques are not yet routinely applied due to existing limitations. Addressing challenges related to CTC epitope expression, cell capture efficiencies, processing of large blood volume, and workflow automation, is essential.
For ctDNA detection technology to be effective, it is crucial to consider that ctDNA accounts only for a small percentage of total cell-free DNA in blood.4 Despite this fact, advancements in ctDNA analysis have improved detection limits, particularly with third-generation sequencing techniques. High-throughput next-generation sequencing (NGS) and digital PCR (dPCR) have emerged as promising technologies for the detection of mutations in peripheral blood samples.13 NGS offers extensive information by simultaneously evaluating multiple genetic and epigenetic aberrations over millions of ctDNA molecules, but it is time-consuming, costly, and lacks long-term monitoring capabilities. In contrast, droplet-based dPCR, an innovative dPCR method based on nanoliter-sized water-in-oil emulsion droplet technology, is particularly suitable for the absolute quantification of nucleic acids, including biomarkers of rare CTC subclones and ctDNA. Droplet-based dPCR excels at absolute quantification, rare mutation detection, copy number variation analysis, DNA methylation analysis, and examination of gene rearrangement across various clinical samples (Figure 2).13 Compared to NGS, droplet-based dPCR is faster, easier to set up, more sensitive, and doesn’t require complex bioinformatics analysis.
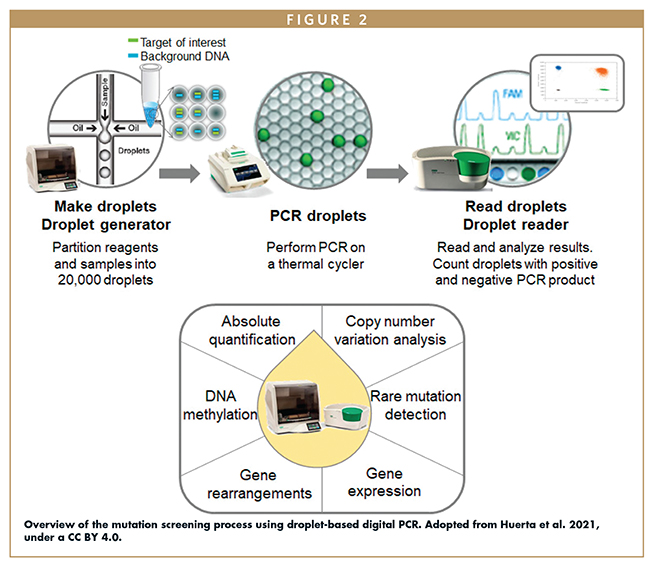
Application of droplet-based dPCR has successfully demonstrated clinical follow-up potential for various cancers through DNA mutation detection using liquid biopsy samples. The droplet-based multiplex dPCR method has been used to detect KRAS mutations in ctDNA of metastatic colorectal cancer patients, showcasing its potential in monitoring therapy response.14 Additionally, this method has also been used to monitor therapy response in stage IV melanoma patients.14 The advancements in PCR-based ctDNA analysis, particularly in droplet-based dPCR methods, have significantly improved the limit of detection and enabled the detection of rare variations.
In summary, integrating data from both CTCs and ctDNA could potentially provide for a more robust evaluation of a tumor’s aggressiveness, potential for metastasis, and overall prognosis. Ongoing research focuses on overcoming current limitations in the field, aiming to improve the specificity, reproducibility, and validation of ctDNA and CTC-based detection systems.2,9,14 Meanwhile, advancements in microfluidics, artificial intelligence, and machine learning for data analysis, and improvements in sequencing technologies aim to enhance the accuracy and reliability of co-analysis. Collaborative efforts between researchers, clinicians, and industry partners are essential for bringing these innovations into routine clinical practice.
LOOKING INTO THE FUTURE
The co-analysis of CTCs and ctDNA represents a paradigm shift in our approach to understanding cancer tumor heterogeneity. Long-term monitoring of phenotypic, genomic, and epigenetic changes in CTCs and ctDNA mutations would provide valuable insights into the effectiveness of therapeutic interventions.1 By correlating genetic alterations detected in ctDNA with captured CTCs characteristics, researchers can establish connections between genomic, phenotypic, and functional changes and gain a better understanding of tumor biology. The potential clinical applications of this approach, from personalized medicine to dynamic treatment monitoring, hold immense promise for improving cancer outcomes by guiding adjustments to treatment strategies. While challenges persist, ongoing research and technological innovations pave the way for a future where the co-analysis of CTCs and ctDNA becomes a routine and invaluable tool in the fight against cancer.
REFERENCES
- Ring A, Nguyen-Sträuli BD, Wicki A, Aceto N. Biology, vulnerabilities and clinical applications of circulating tumour cells. Nat Rev Cancer. 2023;23(2):95-111. doi:10.1038/s41568-022-00536-4.
- Wang X, Wang L, Lin H, et al. Research progress of CTC, ctDNA, and EVs in cancer liquid biopsy. Front Oncol. 2024;14:1303335. doi:10.3389/fonc.2024.1303335.
- van de Stolpe A, Pantel K, Sleijfer S, Terstappen LW, den Toonder JM. Circulating tumor cell isolation and diagnostics: toward routine clinical use. Cancer Res. 2011;71(18):5955-5960. doi:10.1158/0008-5472.CAN-11-1254.
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients.Nat Rev Cancer. 2011;11(6):426-437. doi:10.1038/nrc3066.
- Pessoa LS, Heringer M, Ferrer VP. ctDNA as a cancer biomarker: A broad overview. Crit Rev Oncol Hematol. 2020;155:103109. doi:10.1016/j.critrevonc.2020.103109.
- Bates M, Mohamed BM, Ward MP, et al. Circulating tumour cells: The Good, the Bad and the Ugly. Biochim Biophys Acta Rev Cancer. 2023;1878(2):188863. doi:10.1016/j.bbcan.2023.188863.
- Trujillo B, Wu A, Wetterskog D, Attard G. Blood-based liquid biopsies for prostate cancer: clinical opportunities and challenges. Br J Cancer. 2022;127(8):1394-1402. doi:10.1038/s41416-022-01881-9.
- Yin A, Veerman GDM, van Hasselt JGC, et al. Quantitative modeling of tumor dynamics and development of drug resistance in non-small cell lung cancer patients treated with erlotinib. CPT Pharmacometrics Syst Pharmacol. 2024; 00: 1-12. doi:10.1002/psp4.13105.
- Neumann MHD, Bender S, Krahn T, Schlange T. ctDNA and CTCs in Liquid Biopsy – Current Status and Where We Need to Progress. Comput Struct Biotechnol J. 2018;16:190-195. doi:10.1016/j.csbj.2018.05.002.
- Dual isolation and profiling of circulating tumor cells and circulating tumor DNA from a single liquid biopsy sample using the Genesis Isolation System with Celselect Slides and Droplet Digital PCR. Bulletin 3599. Available from: https://www.bio-rad.com/sites/default/files/2023-11/Bulletin_3599.pdf.
- Visal TH, den Hollander P, Cristofanilli M, Mani SA. Circulating tumour cells in the -omics era: how far are we from achieving the ‘singularity’?. Br J Cancer. 2022;127(2):173-184. doi:10.1038/s41416-022-01768-9.
- Circulating tumor cell (CTC) enumeration overcomes limitations of other blood-based cancer detection methods. Bulletin 3267. Available from: https://www.bio-rad.com/sites/default/files/2021-12/DOC007-R10-Enumeration-and-Enrichment-App-Note-24NOV2021.pdf.
- Huerta M, Roselló S, Sabater L, et al. Circulating Tumor DNA Detection by Digital-Droplet PCR in Pancreatic Ductal Adenocarcinoma: A Systematic Review. Cancers (Basel). 2021;13(5):994. doi:10.3390/cancers13050994.
- K. Yi, X. Wang, S. K. Filippov, et al. Emerging ctDNA detection strategies in clinical cancer theranostics. Smart Med. 2023, 2(4), e20230031. doi: 10.1002/SMMD.20230031.

Dr. Yoon-Tae Kang is a biomedical researcher and Staff Scientist at Life Science Group, Bio-Rad Laboratories, who has developed novel microfluidic/nanofluidic platforms and systems for diagnosing cancer and neurodegenerative diseases using minimal sample volumes in less invasive methods (called liquid biopsy). His major focus at Bio-Rad is to develop a market where Bio-Rad’s flagship product lines can facilitate biological findings and clinical implications through various downstream analysis and brand-new approaches in sample processing. Prior to joining Bio-Rad, he was a Biointerfaces Institute (BI) Innovation Fellow and a senior scientist at University of Michigan, who was responsible for developing cutting-edge microsystems for circulating tumor cells/small-extracellular vesicles (sEV) isolation and characterization. He is a first/corresponding author of 18 peer-reviewed journal papers, guest editor of Applied Science, and reviewer of high-impact peer-reviewed journals from Wiley, ACS, Royal Society of Chemistry, and Science Publishing Group.

Dr. Abiodun ‘Abi’ Bodunrin is an Application Scientist in the Market Development and Applications team at the Life Science Group, Bio-Rad Laboratories. He develops products and tools, gathers data, and authors high impact scientific content for applications in Oncology research, pathogen detection in wastewater, and infectious diseases. He also collaborates with Marketing, Business Development, and R&D on new product development process. He graduated with a PhD in Biochemistry from the University of Houston, where he worked on characterizing Universal Stress Proteins (USPs) implicated in Dormancy regulation in a Gram-Positive Bacteria under the mentorship of Dr. William Widger. He earned his Master of Science in Biotechnology from West Virginia State University. He is a first/corresponding author of 11 peer-reviewed publications, technical application notes, and protocols.

Dr. Joby Chesnick is a Senior Segment Marketing Manager in the Digital Biology Group at Bio-Rad Laboratories, where she manages marketing and communications for the Single Cell product portfolio. She earned her BS and MS in Biology at Western Illinois University, followed by a PhD in Plant Biology at Texas A&M University. She went on to receive a National Science Foundation Post-Doctoral Fellowship in Plant Biology while at the University of Washington, where she characterized the chloroplast genome of an algal (dinoflagellate & diatom) endosymbiosis to elucidate the mechanisms of early organelle biogenesis. She has since held a variety of positions in both the biotech and molecular diagnostics industries, earned her MBA at the University of Wisconsin, and most recently joined Bio-Rad Laboratories in 2022 in her present role.
Total Page Views: 2669