Issue:May 2016
PROTEIN PHOSPHORYLATION - New Pathways, New Tools: Phosphohistidine Signaling in Mammalian Cells
HISTIDINE PHOSPHORYLATION: A THERAPEUTICALLY IMPORTANT PATHWAY?
Because protein phosphorylation regulates a wide array of cellular processes, many drugs and drug candidates act on kinases and phosphatases to modulate post-translational modifications (PTMs). As of April 2015, the United States Food and Drug Administration (FDA) had approved 28 therapeutic protein kinase inhibitors, the majority of which are being used to treat cancers. Most drug discovery programs are focused on tyrosine, serine, and threonine kinases; however, protein phosphorylation is known to occur on nine different amino acids. Systems in which multiple amino acids are phosphorylated and dephosphorylated may have an evolutionary advantage – regulation by multiple kinases and phosphatases may confer more opportunities to respond to environmental cues and, thus, increase capacity to dynamically tune a response.
Could the phosphorylation of histidine emerge as a therapeutically important pathway in mammals? The phosphorylation of histidine has not been widely studied in mammalian cells, despite its discovery in bovine liver mitochondria.1 Histidine phosphorylation is well-studied in bacteria and is part of a “two-component” signaling pathway used for chemotaxis and transcription (Figure 1). In prokaryotes, this signaling system is a phosphorelay system typically consisting of two types of proteins: a membrane-bound histidine kinase (HK) and a response regulator (RR). This system is activated when an environmental signal causes a membrane-associated HK to be auto-phosphorylated on a conserved histidine residue. This is followed by the transfer of the phosphate to a cytoplasmic RR protein on a conserved aspartate residue. The phosphorylated RR protein can then function as a transcription factor and activate genes associated with range of cellular processes, such as chemotaxis, stress response, sporulation, antibiotic resistance, etc.2
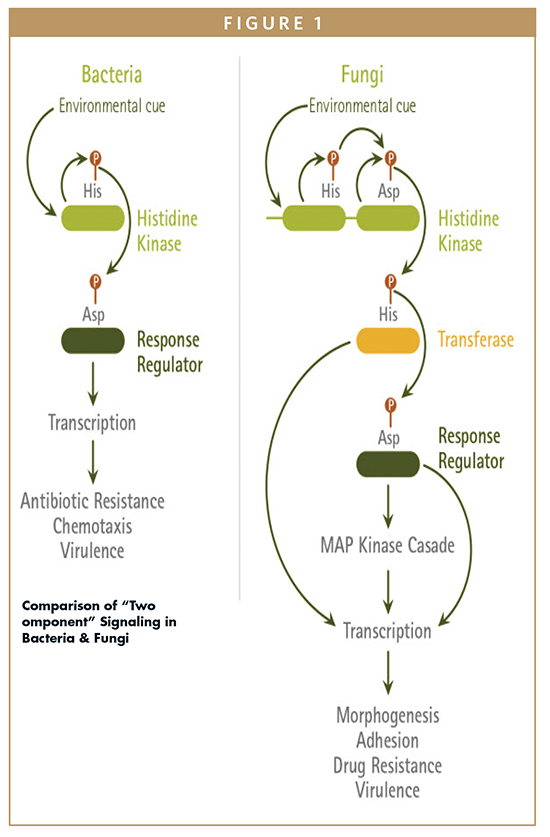
Fungi also possess a similar two-component system, which is more complex than the bacterial system. In the fungal system, the HK and RR proteins are present, as well as a third protein, a histidine phosphotransferase (HPt). In yeast, this protein is known as Phosphorelay intermediate protein Ypd1. The main function of the Ypd1 protein is to facilitate the transfer of phosphate from the histidine kinase to response regulator proteins.3 Another key difference between the bacterial and fungal two-component systems is the number of phosphorylation events that occur (Figure 1). In fungal systems, there are four phosphorylation events that happen on the three proteins. First, the HK protein is auto-phosphorylated on a histidine present in its histidine kinase domain. This event is followed by a transfer of the phosphate group to an aspartate on the receiver domain of the HK protein. A third phosphotransfer event occurs in which the phosphate group is transferred to the histidine phosphotransferase domain on the Ypd1 protein. Finally, a fourth transfer occurs in which the phosphate group is transferred to an aspartate on the RR protein. This can result in either the activation or downregulation of specific sets of genes, depending on the system and the specific sets of proteins involved.
While not as well-characterized, histidine phosphorylation has been detected in eukaryotic systems.4-5 Recent evidence suggests phosphohistidine signaling is involved in the regulation of key cellular processes in mammalian cells.5 In support of this assertion, two mammalian histidine protein kinases have been identified. These protein kinases are known as NME1 (Nucleoside diphosphate kinase A or Nm23-H1) and NME2 (Nucleoside diphosphate kinase B or Nm23-H2). Recent data are consistent with roles for these enzymes in a variety of cellular processes including mammalian signal transduction,6-8 cancer biology,9,10 differentiation, development, apoptosis, cytokinesis, and dynamin-mediated endocytosis.11,12
Some phosphohistine-containing proteins that are substrates of NME1 and NME2 have been recently identified. One of these is SK4, a calcium-activated potassium channel expressed in mammalian blood vessels, whose expression is correlated with the proliferation of endothelial cells in cardiovascular disease.13 SK4 can be reversibly phosphorylated at His358, enabling the fine regulation of vascular cell proliferation.14 Another study identified the calcium channel, TRPV5, as a protein regulated by histidine phosphorylation by NME1.6 When phosphorylated at His711, the TRPV5 channel shows increased activity, as shown by patch clamp experiments. More calcium enters TRPV5-expressing cells in NME1 wild-type background compared to cells in which NME1 has been knocked down. The study’s authors, however, cite “technical problems with our assays, as well as reagents that are available to detect histidine phosphorylated proteins” as reasons for not being able to collect more detailed information regarding the histidinemediated regulation of TRPV5.6
OBSTACLES TO DETECTING HISTIDINE PHOSPHORYLATION
One reason detecting histidine phosphorylation has been so difficult is that the modification is thermodynamically less stable than phosphorylation of serine, threonine or tyrosine. Compared to phosphorylation of hydroxyl groups, which exhibit ΔG of hydrolysis ranging from -6.5 to -9.5 kcal/mol, the phosphorylation of histidine has a ΔG of hydrolysis ranging from -12 to -14 kcal/mol4. Furthermore, under the acidic conditions in which phosphoproteins are frequently purified, the kinetic stability of phosphohistidine is lower than that of other modifications.4 Thus, histidines often become dephosphorylated during preparation for mass spectrometry and acidic staining of polyacrylamide gels, and other manipulations. Phosphoserine and phosphothreonine are sometimes purified using chromatography in which the stationary phase is conjugated to metal ions; however, because both phosphorylated and unphosphorylated histidine can bind to many metal ions, this method has been less fruitful for phosphohistidine detection.
Progress in understanding the cellular function of phosphohistidine has been further hampered by the lack of specific antibody detection reagents for this modification. Creation of these reagents using phosphohistidine peptides as an immunogen has proven challenging due to the instability of phosphoramidate bonds resulting in the destruction of the immunogen before a strong immune response can occur. The effort was even called “devilishly hard” by Kee and Muir.15
ANTIBODIES AGAINST PHOSPHOHISTIDINE: THE GOOD, THE BETTER & THE BEST YET
Recently, pan-phosphohistidine antibodies were generated using nonhydrolyzable synthetic analogs of phosphohistidine.16,17 For the first of these antibodies, peptides containing phosphohistidine were recognized with only ten-fold higher sensitivity than peptides containing phosphotyrosine.16 The cross-reactivity with phosphotyrosine was even greater when assessed using bovine serum albumin-conjugated (BSA) phosphotyrosine and phosphohistidine.17 The second pan-phosphohistidine antibody showed less, but still significant, crossreactivity with phosphotyrosine in the context of a bovine serum albumin conjugate.17
Additionally, and even more significantly, these antibodies were unable to distinguish between the N-1 and N-3 phosphoisomers of histidine. One unique feature of histidine is that phosphohistidine can be phosphorylated at either the N-1 or N-3 nitrogen of the imidazole ring (Figure 2). This is because at physiological pH, the imidazole ring of histidine can have its proton at either nitrogen. Thus, in biological systems, two tautomers exist, resulting from phosphorylation at either N1 (1-pHis) or N3 (3-pHis) positions (Figure 2).
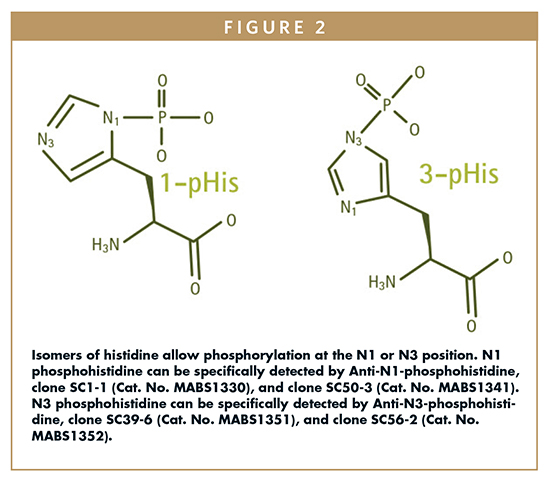
Both the crossreactivity with phosphotyrosine and the inability to distinguish between phosphoisomers present a challenge to researchers wishing to detect phosphohistidine in proteomics studies of eukaryotes more complex than yeast. To better understand the role of phosphohistidine signaling in mammalian systems, antibodies that have the following properties are needed:
-Sequence-independent detection of phosphohistidine
-Isomer-specific antibodies that reliably discriminate between 1-pHis or 3-pHis
-No cross reactivity with phosphotyrosine
-The ability to work in a variety of applications, such as Western blotting, immunofluorescence and immunoaffinity purification
With these specific goals, a team of Salk Institute researchers, led by Tony Hunter, engaged in a project to generate a set of monoclonal antibodies with these properties. Their success in generating these highly specific and isoform-specific monoclonal antibodies was recently reported in Cell.18
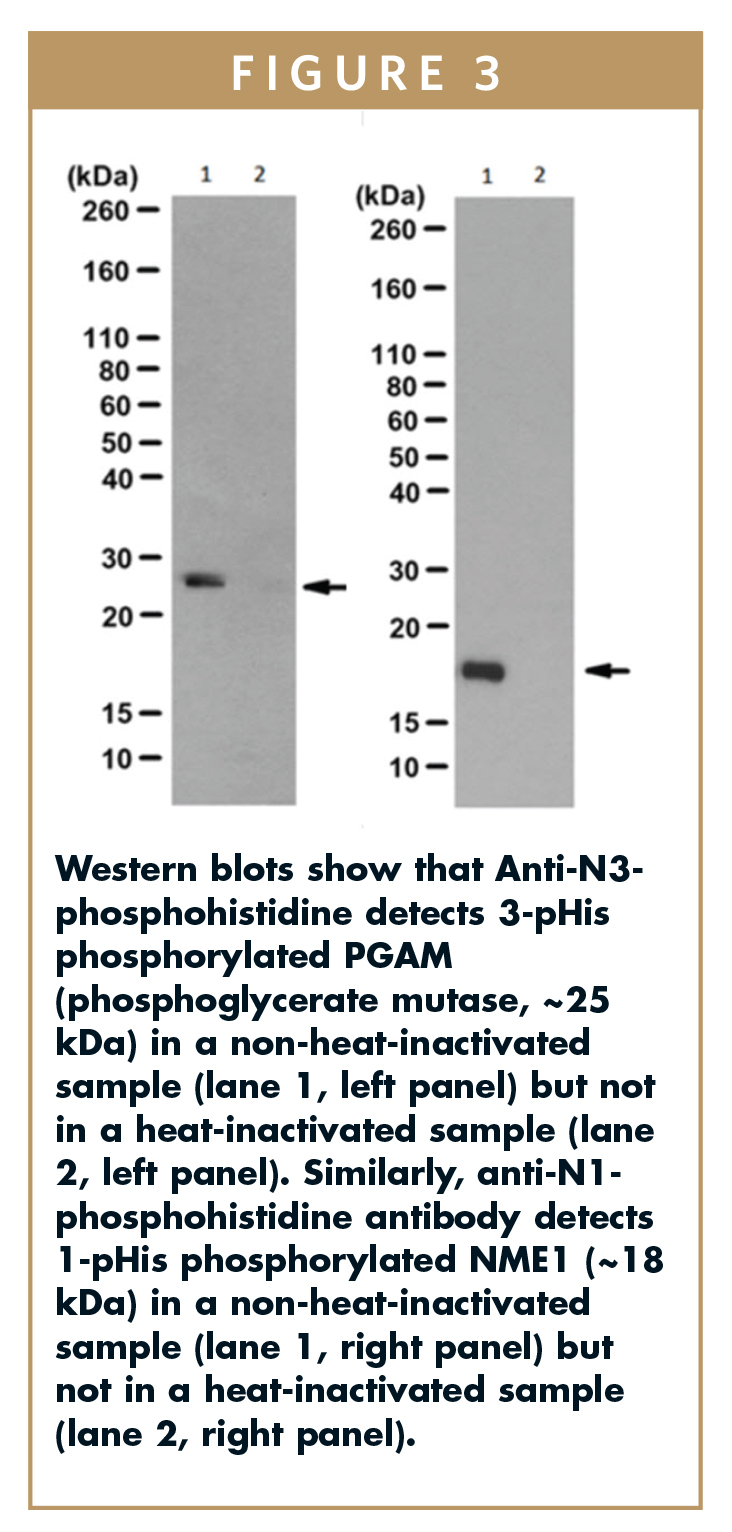
To get around the issue with the stability of the phosphoramidate bond, the Hunter lab created degenerate peptide libraries that incorporated nonhydrolyzable phosphoryl-triazolylalanine (pTza) histidine analogs into the peptide sequence. For the generation of isoform-specific antibodies, two separate degenerate peptide libraries containing either 1-pTza or 3-pTza were used for immunizations. The resulting antisera were initially screened using dot blots against the immunizing peptides to identify isomer-specific sera. Sera with the right specificity profile were further characterized by Western blotting using proteins phosphorylated in vitro using NME1/NME2 (generates 1-pHis modified proteins) or PGAM (generates 3-pHis modified proteins), with heat-induced dephosphorylation as a negative control (Figure 3). Anti-N3-phosphohistidine detected 3-pHis phosphorylated PGAM (phosphoglycerate mutase, ~25 kDa) in a non-heat-inactivated sample but not in a heat-inactivated sample. Similarly, anti-N1-phosphohistidine antibody detected 1-pHis phosphorylated NME1 (~18 kDa) in a non-heat-inactivated sample but not in a heat-inactivated sample.
Hybridomas were generated and screened to yield multiple isoform-specific monoclonal antibodies.To demonstrate that the resulting monoclonals detected pHis in a sequence-independent fashion, each of these monoclonal antibodies was screened using pTza peptides of a defined sequence. This was followed by screening with cell lysates to demonstrate that these antibodies are capable of detecting endogenous pHis proteins. The specificity for phosphohistidine and lack of cross reactivity to pTyr was also evaluated using both pTyr peptides and cell lysates. Neither the 1-pHis nor the 3-pHis monoclonal antibodies generated showed any cross reactivity to pTyr.
WHAT THE NEW PHIS ANTIBODIES REVEAL
In their Cell publication, the Hunter lab presented data from both immunofluorescence and immunoaffinity purification experiments.18 Using the monoclonal antibodies developed for this study, they evaluated the biological role of the phosphohistidine modification in a manner not previously possible and reported some interesting findings relative to the two isoforms of phosphohistidine. Specifically, immunofluorescence using 1-pHis antibodies was localized to the outer membrane of phagosomes. Phagosomes are vesicles that are formed when the cell membrane closes completely around a particle, for the purposes of nutrition or cellular defense; this pHis localization pattern is consistent with the previous finding that histidine kinases may regulate membrane trafficking.11 In contrast, immunostaining with 3-pHis antibodies suggested involvement in the cell cycle with the N-3 phosphoisomer associated with centrosomes, spindle poles, and the midbodies of cells in late telophase.
To identify specific proteins containing the pHis modification, immunoaffinity purification was performed, followed by mass spectrometry. This work identified 280 different proteins containing the 1-pHis modification and 156 proteins with the 3-pHis modification. This diversity of proteins that are uniquely associated with either the 1-pHis or 3-pHis isoform suggests a role for phosphohistidine in a variety of cellular processes. Mass spectrometry analysis of the 3-pHis proteins was consistent with the immunofluorescence data, with several of the 3-pHis proteins identified being associated with the cell cycle.
The work of the Hunter lab has resulted in the generation of an important set of reagents. The existence of these phosphohistidine monoclonal antibodies will pave the way for more detailed studies of the role of histidine phosphorylation in mammalian systems. Studies using these reagents can help better define the emerging roles of phosphohistidine in areas such as signal transduction, cancer, differentiation, development, and apoptosis.
Making these reagents readily available to the scientific community will help drive this understanding. MilliporeSigma has licensed the Hunter lab’s phosphohistidine antibodies to make them easily available to the research community. It is expected that the broad availability of these reagents will open up the study of histidine phosphorylation to a broad segment of researchers, and uncover new paradigms in cellular signaling.
THE FUTURE OF PTM RESEARCH
The methods by which the new phosphohistidine antibodies were developed may guide researchers to developing antibodies to other post-translational modifications and “challenging” epitopes. After all, there are other amino acids that can be phosphorylated (albeit unstably) by protein kinases: cysteine, glutamate, aspartate, and lysine. Most recently, chemists at Xiamen University reported developing a structural analog of phosphoarginine, using the same general synthetic scheme as used by the Hunter lab, with the goal of using this analog as a hapten immunogen for raising antiphosphoarginine antibodies.19 We do not yet fully understand the importance of phosphorylation of these other amino acids is in eukaryotes; however, the development analytical methods adapted for neutral or basic pH, combined with the development of novel antibodies, can help answer that question. Affinity-based enrichment of proteome samples using specific phosphoamino acid antibodies can then be combined with advanced mass spectrometry methods and analysis software that are optimized to fragment and identify PTM-bearing peptides, further aiding the discovery of new PTM sites, kinase substrates, and sophisticated cell signaling networks.20
Once studies reveal a critical number of therapeutically promising targets bearing a particular phosphoamino acid, one would expect a subsequent emergence of drug discovery and development programs targeting the regulation of this PTM. In the case of histidine phosphorylation, several small molecule inhibitors of bacterial histidine kinases have been developed as antibiotics.21 Further study of mammalian histidine kinases may reveal the utility of these scaffolds as starting points for targeting therapeutically relevant pathways in humans.
REFERENCES
1. Boyer PD, et al. Identification of phosphohistidine in digests from a probable intermediate of oxidative phosphorylation. J Biol Chem. 1962; 237:PC3306-PC3308. [http://www.ncbi.nlm.nih.gov/pubmed/14014715].
2. Mascher T. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol Lett. 2006 Nov;264(2):133-44. [http://www.ncbi.nlm.nih.gov/pubmed/17064367].
3. Fassler JS and West AH. Histidine phosphotransfer proteins in fungal two-component signal transduction pathways. Eukaryot Cell. 2013;12(8):1052-60 [http://www.ncbi.nlm.nih.gov/pubmed/23771905].
4. Besant PG, Attwood PV. Detection and analysis of protein histidine phosphorylation. Mol Cell Biochem. 2009 Sep;329(1-2):93-106.
5. Attwood PV. Histidine kinases from bacteria to humans. Biochem. Soc. Trans. 2013;411023-1028. [http://www.ncbi.nlm.nih.gov/pubmed/23863173].
6. Cai X, Srivastava S, Surindran S, et al. Regulation of the epithelial Ca²⁺ channel TRPV5 by reversible histidine phosphorylation mediated by NDPK-B and PHPT1. Mol Biol Cell. 2014;25(8):1244-50. [http://www.ncbi.nlm.nih.gov/pubmed/24523290].
7. Hartsough MT, Morrison DK, Salerno M, et al. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem. 2002;277(35):32389-99. [http://www.ncbi.nlm.nih.gov/pubmed/12105213].
8. Feng Y, Gross S, Wolf NM, et al. Nucleoside diphosphate kinase B regulates angiogenesis through modulation of vascular endothelial growth factor receptor type 2 and endothelial adherens junction proteins. Arterioscler Thromb Vasc Biol. 2014 Oct;34(10):2292-300. [http://www.ncbi.nlm.nih.gov/pubmed/25147336].
9. Thakur RK, Yadav VK, Kumar P, et al. Mechanisms of non-metastatic 2 (NME2)-mediated control of metastasis across tumor types. Naunyn Schmiedebergs Arch Pharmacol. 2011;384(4-5):397-406. [http://www.ncbi.nlm.nih.gov/pubmed/21556888].
10. Tso PH, Wang Y, Yung LY, et al. RGS19 inhibits Ras signaling through Nm23H1/2-mediated phosphorylation of the kinase suppressor of Ras. Cell Signal. 2013;25(5):1064-74. [http://www.ncbi.nlm.nih.gov/pubmed/23416464].
11. Boissan M, Montagnac G, Shen Q, et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science. 2014;344(6191):1510-5. [http://www.ncbi.nlm.nih.gov/pubmed/24970086].
12. Conery AR, Conery AR, Sever S, Harlow E. Nucleoside diphosphate kinase Nm23-H1 regulates chromosomal stability by activating the GTPase dynamin during cytokinesis. Proc Natl Acad Sci USA. 2010;107(35):15461-6. [http://www.ncbi.nlm.nih.gov/pubmed/20713695].
13. Srivastava S, Zhdanova O, Di L, et al. Protein histidine phosphatase 1 negatively regulates CD4 T cells by inhibiting the K+ channel KCa3.1. Proc. Natl. Acad. Sci. USA. 2008;105:14442-14446. [http://www.ncbi.nlm.nih.gov/pubmed/18796614].
14. Zhou XB, Feng YX, Sun Q, et al. Nucleoside diphosphate kinase B-activated intermediate conductance potassium channels are critical for neointima formation in mouse carotid arteries. Arterioscler Thromb Vasc Biol. 2015;35:1852-1861. [http://www.ncbi.nlm.nih.gov/pubmed/26088577].
15. Kee JM, Muir TW. Chasing phosphohistidine, an elusive sibling in the phosphoamino acid family. ACS Chem Biol. 2012 Jan 20;7(1):44-51.
16. Kee JM, Oslund RC, Perlman DH, Muir TW. A pan-specific antibody for direct detection of protein histidine phosphorylation. Nat Chem Biol. 2013;9(7):416-21 [http://www.ncbi.nlm.nih.gov/pubmed/23708076].
17. Kee JM, Oslund RC, Couvillon AD, Muir TW. A second-generation phosphohistidine analog for production of phosphohistidine antibodies. Org Lett. 2015;17(2):187-9 [http://www.ncbi.nlm.nih.gov/pubmed/25531910].
18. Fuhs SR, Meisenhelder J, Aslanian A, Ma L, et al. Monoclonal 1- and 3-Phosphohistidine Antibodies: New Tools to Study Histidine Phosphorylation. Cell. 2015;162(1):198-210. [http://www.ncbi.nlm.nih.gov/pubmed/26140597].
19. Ouyang H, Fu C, Fu S et al. Development of a stable phosphoarginine analog for producing phosphoarginine antibodies. Org Biomol Chem. 2016 Feb 2;14(6):1925-9 [http://www.ncbi.nlm.nih.gov/pubmed/26750450].
20. Oslund RC, Kee JM, Couvillon AD, et al. A phosphohistidine proteomics strategy based on elucidation of a unique gas-phase phosphopeptide fragmentation mechanism. J Am Chem Soc. 2014 Sep 17;136(37):12899-911. [http://www.ncbi.nlm.nih.gov/pubmed/25156620].
21. Bem AE, Velikova N, Pellicer MT, Baarlen Pv, Marina A, Wells JM. Bacterial histidine kinases as novel antibacterial drug targets. ACS Chem Biol. 2015 Jan 16;10(1):213-24. [http://www.ncbi.nlm.nih.gov/pubmed/25436989].
To view this issue and all back issues online, please visit www.drug-dev.com.
B I O G R A P H I E

Xiaoyi (Jeff) Xu is a Senior Product Manager in the Target Specific Reagents group at MilliporeSigma, the Life Sciences business of Merck KGaA, Darmstadt, Germany, and manages signaling and immunology reagents. A former research scientist in the infectious disease field, he has over 10 years of experience in developing products for the life sciences research market, ranging from instrumentation to research consumables. He earned his MBS from Keck Graduate Institute of Claremont Colleges, focusing on business of bioscience, and his BS in Physiological Science from UCLA.

Dr. Chandreyee Das is a Senior Content Development Specialist at MilliporeSigma, researching trends and technologies in the preparation and analysis of proteins and small molecules in complex samples. She earned her PhD in Chemistry and Chemical Biology at the University of California, San Francisco, studying the structural determinants of RNAprotein interactions, and was a Ruth L. Kirschstein Postdoctoral Fellow in the department of Molecular Oncology at the Dana Farber Cancer Institute.

Dr. Michael R. Sturges served as a Senior Product Manager at MilliporeSigma from 2009 to 2015. He is currently at SGIDNA, a Synthetic Genomics company. For the past 14 years, he has worked in the bioscience industry in marketing and product development. This experience is complemented by more than 12 years of hands-on scientific research at both academic and commercial organizations. Dr. Sturges earned his PhD at the University of California, Santa Cruz, and completed his postdoctoral studies in the Department of Pediatrics in the laboratory of Dr. Charles Roberts, Jr. at Oregon Health and Sciences University. Dr. Sturges is currently applying his experience and knowledge of genomics, proteomics, and molecular biology to drive the development and commercialization of novel technologies that enable research and discovery.
Total Page Views: 4087

















